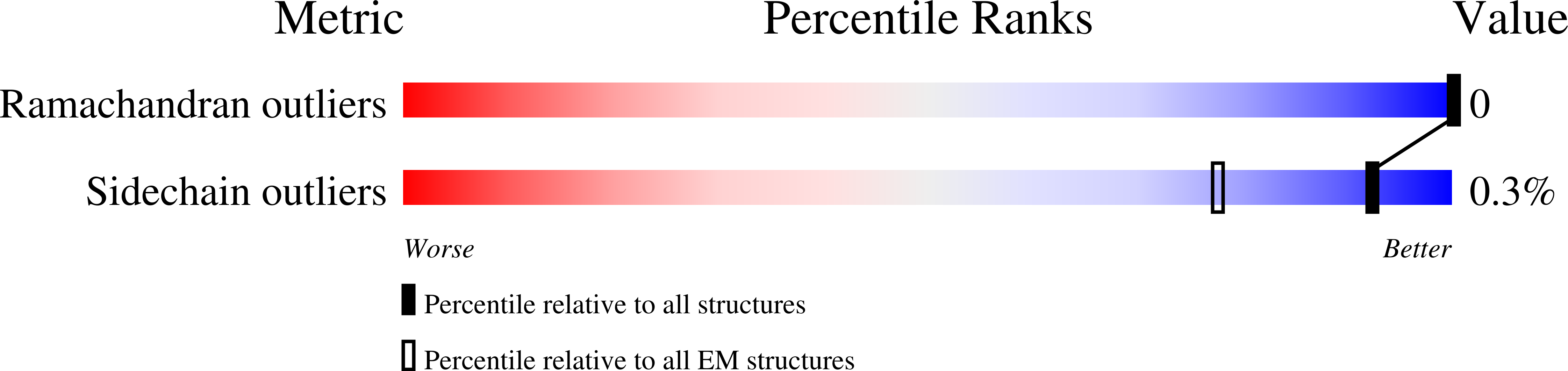
Deposition Date
2023-01-20
Release Date
2023-05-10
Last Version Date
2025-06-04
Entry Detail
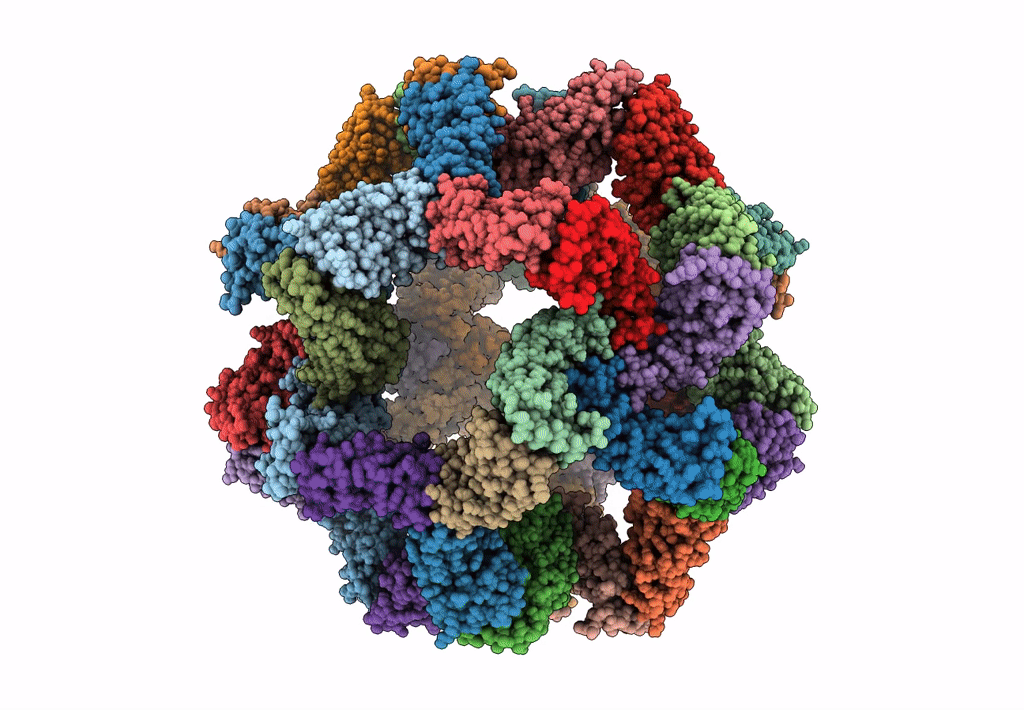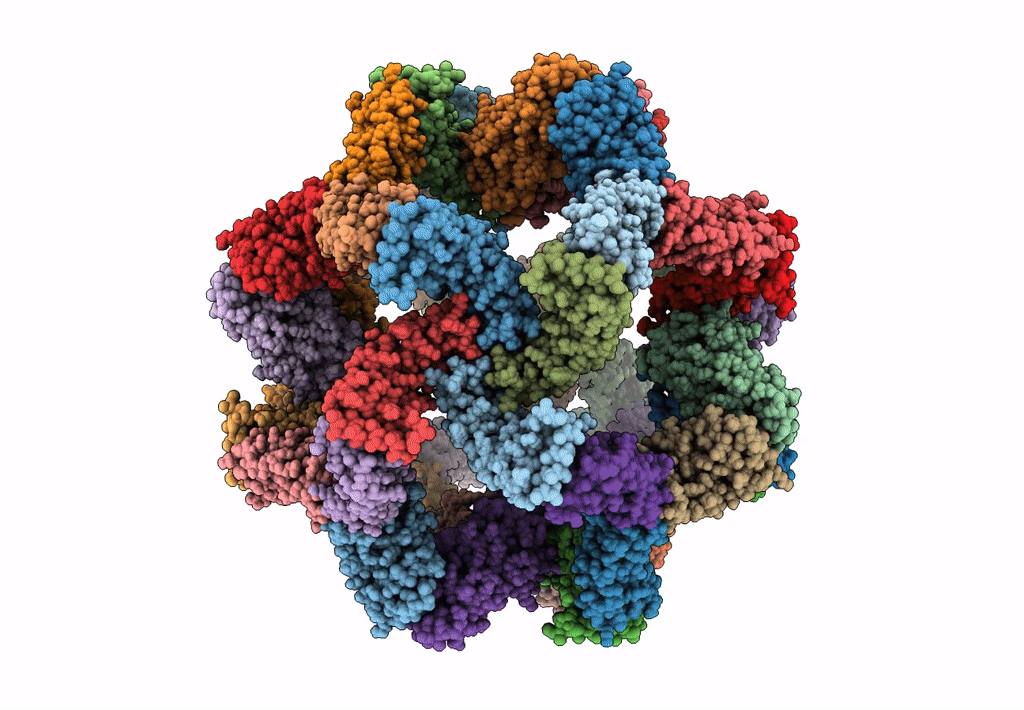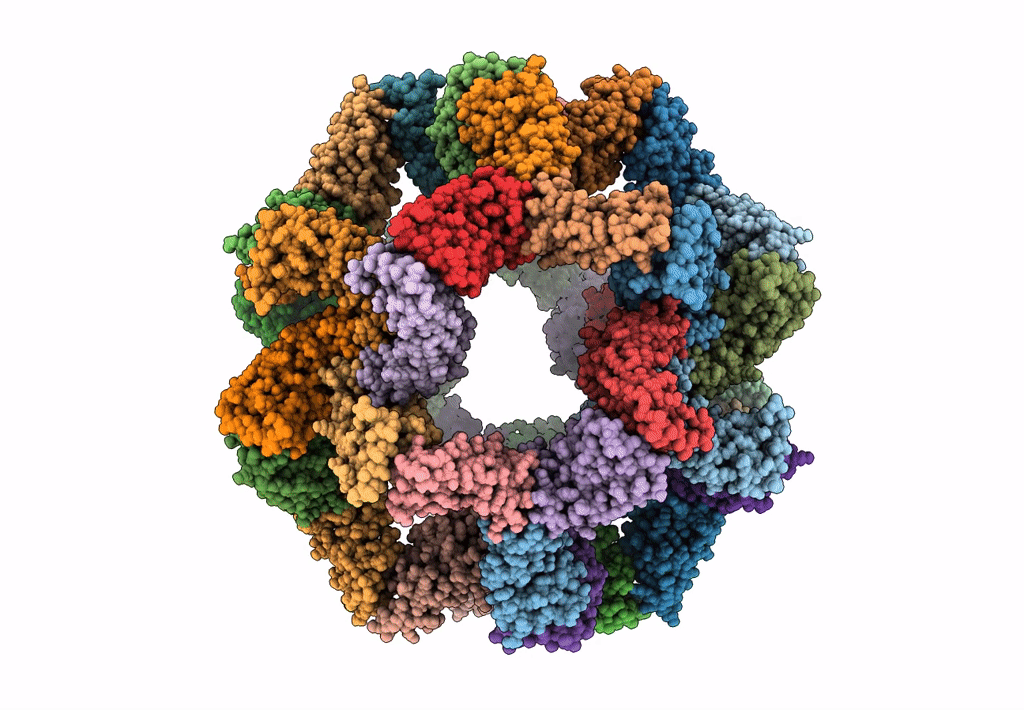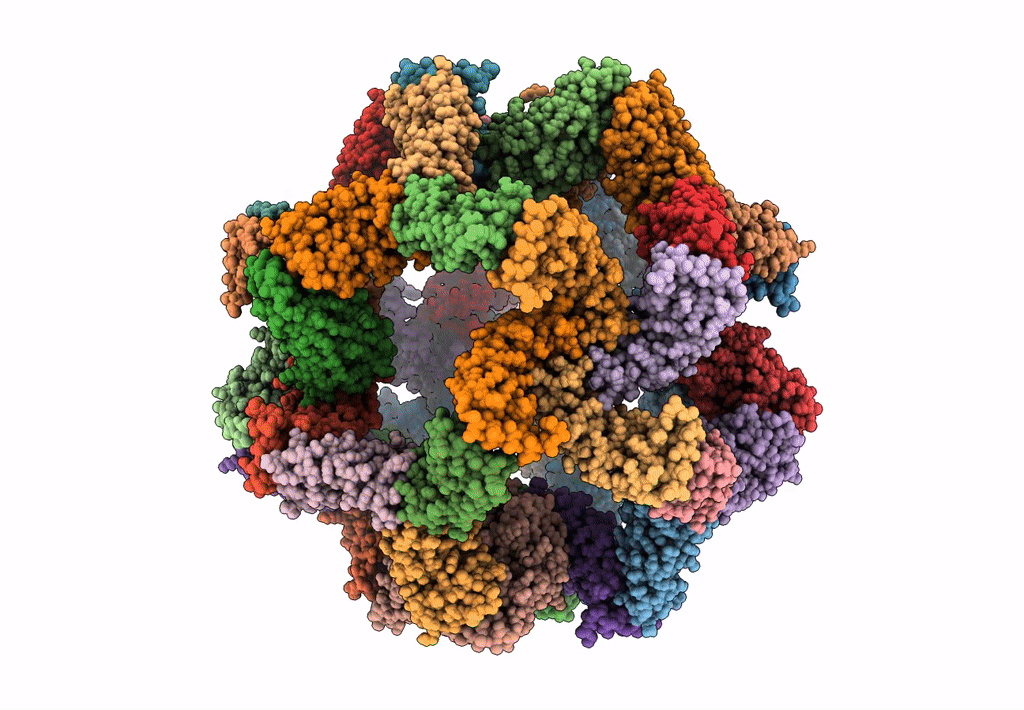
PDB ID:
8FWD
Keywords:
Title:
Fast and versatile sequence- independent protein docking for nanomaterials design using RPXDock
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.67 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE