Deposition Date
2022-10-28
Release Date
2024-02-14
Last Version Date
2024-05-08
Entry Detail
PDB ID:
8EYP
Keywords:
Title:
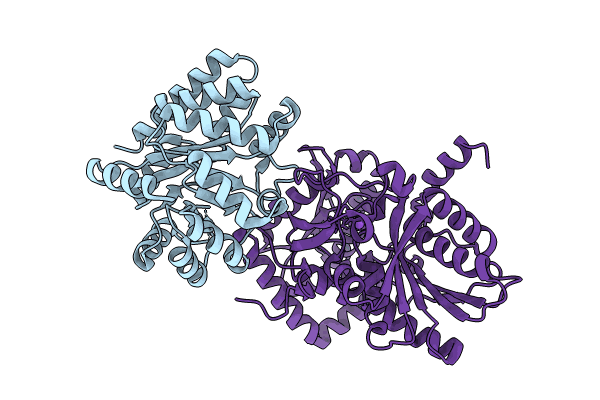
Joint X-ray/neutron structure of Salmonella typhimurium tryptophan synthase internal aldimine from microgravity-grown crystal
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
R-Value Free:
['0.19
R-Value Work:
['0.18
R-Value Observed:
['?', '?'].00
Space Group:
C 1 2 1