Deposition Date
2022-08-09
Release Date
2022-10-26
Last Version Date
2023-10-18
Entry Detail
PDB ID:
8E0F
Keywords:
Title:
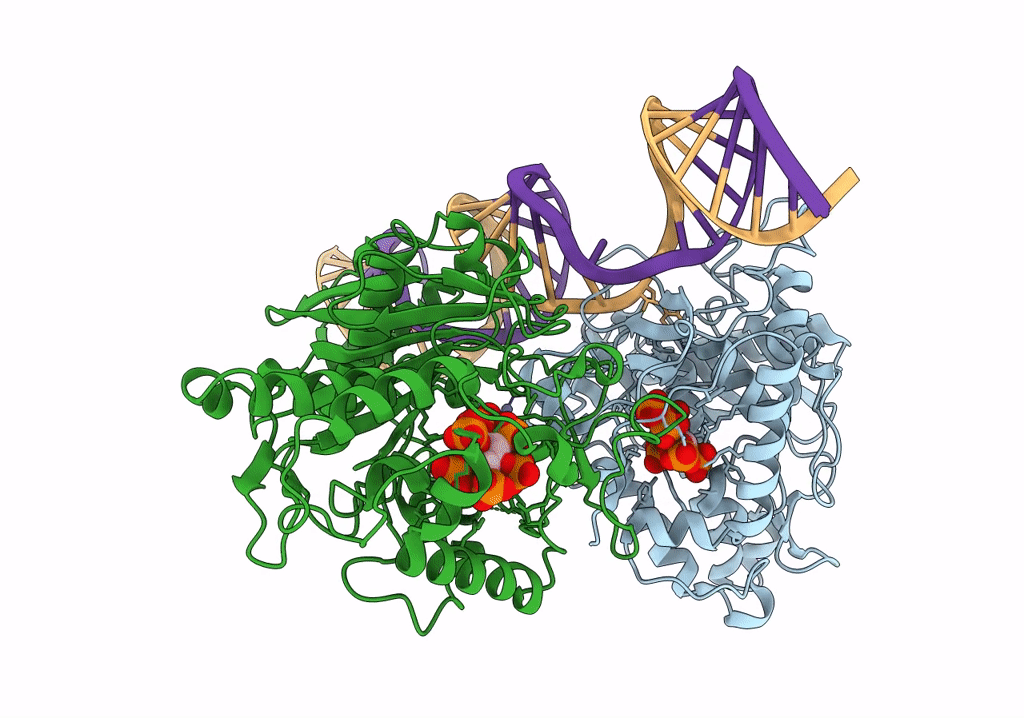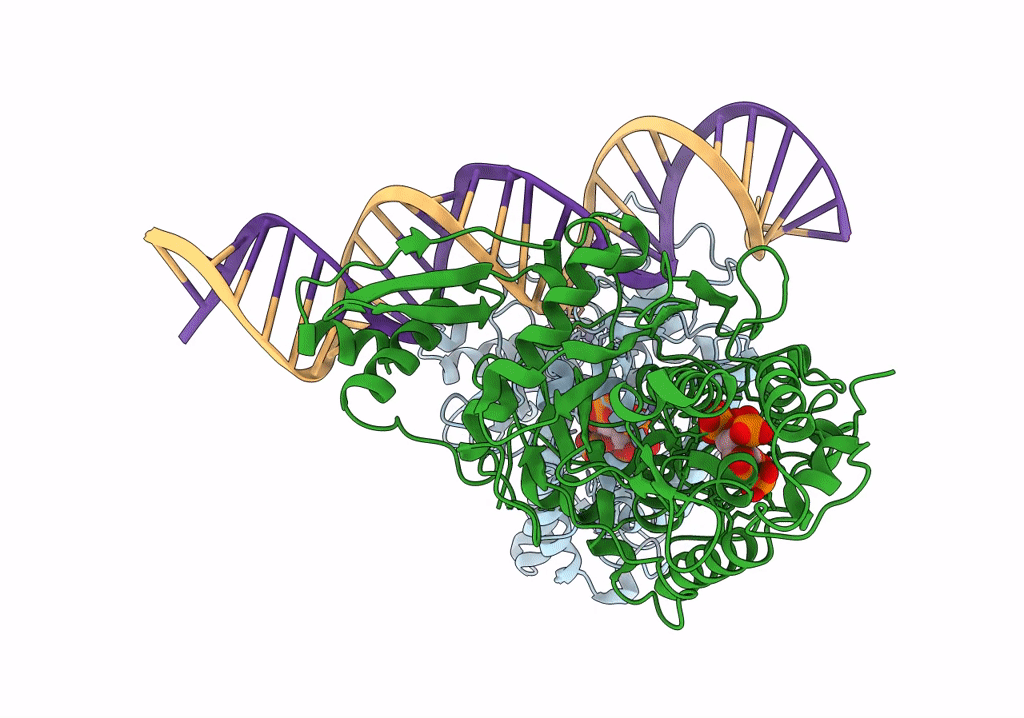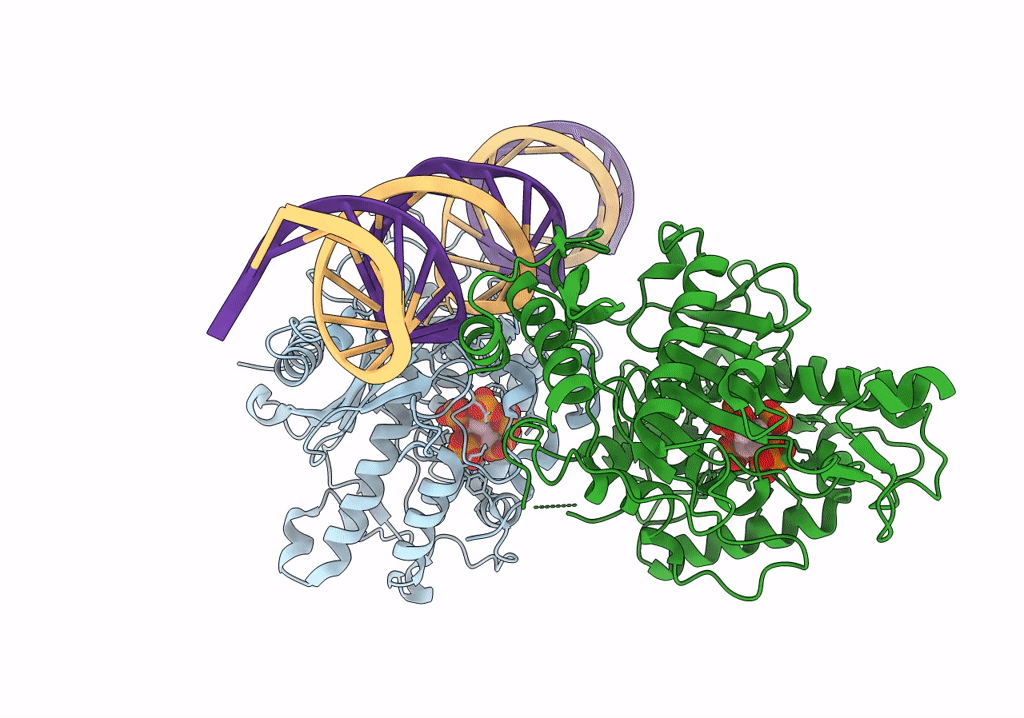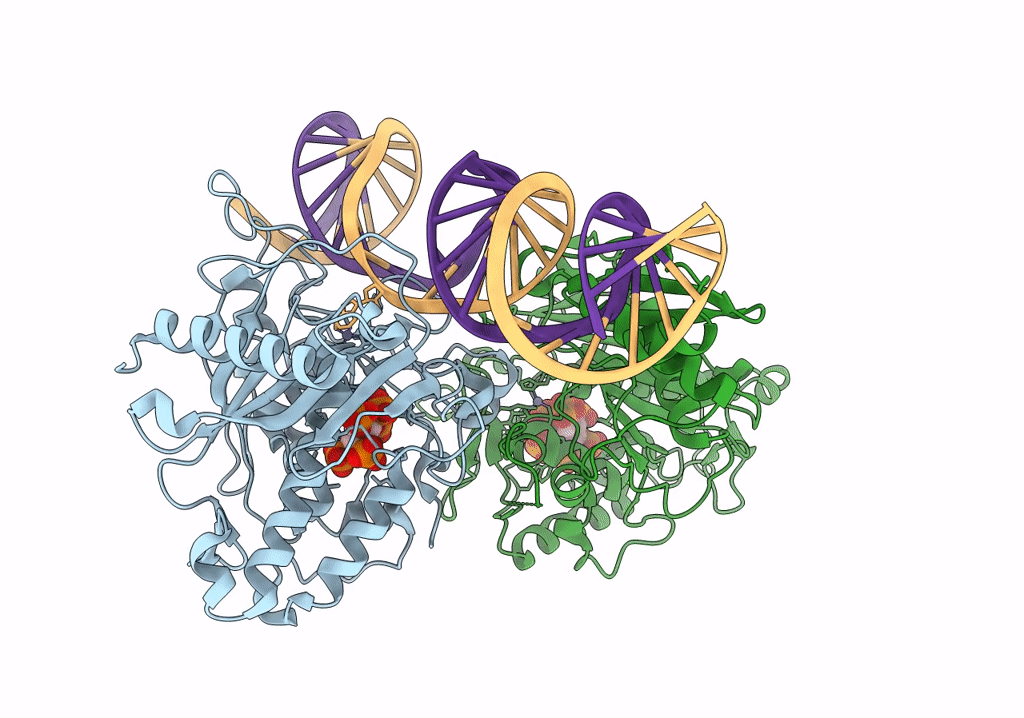
Human Adenosine Deaminase Acting on dsRNA (ADAR2-RD) bound to dsRNA containing a G-G pair adjacent to the target site
Biological Source:
Source Organism:
Homo sapiens (Taxon ID: 9606)
Host Organism:
Method Details:
Experimental Method:
Resolution:
2.70 Å
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
C 1 2 1


