Deposition Date
2022-07-30
Release Date
2022-11-30
Last Version Date
2023-10-25
Entry Detail
PDB ID:
8DVY
Keywords:
Title:
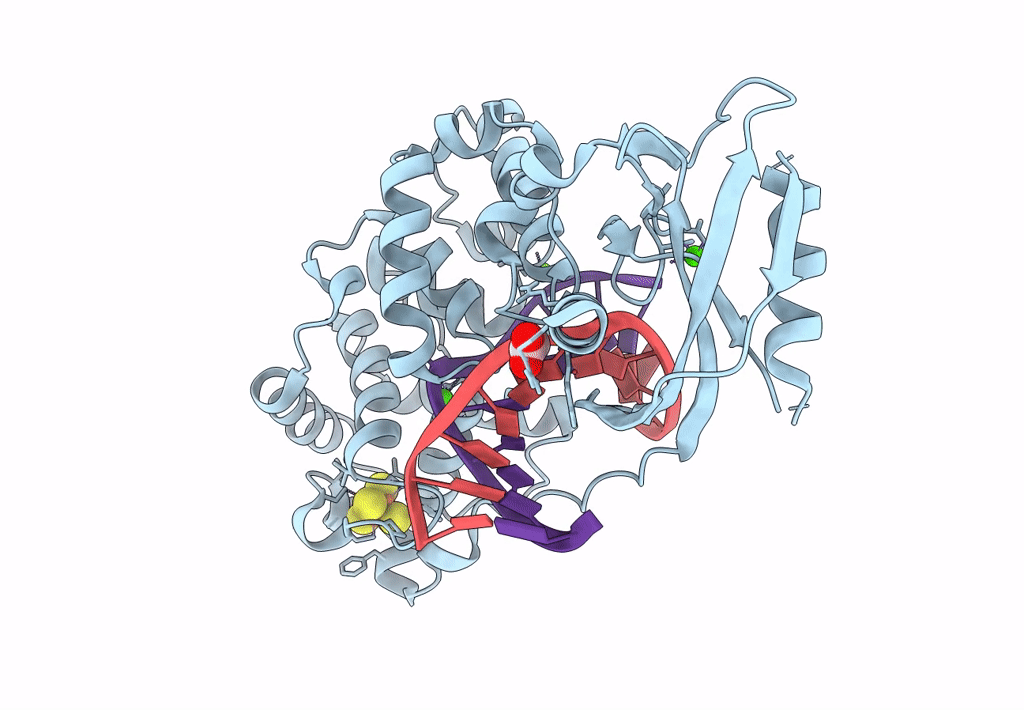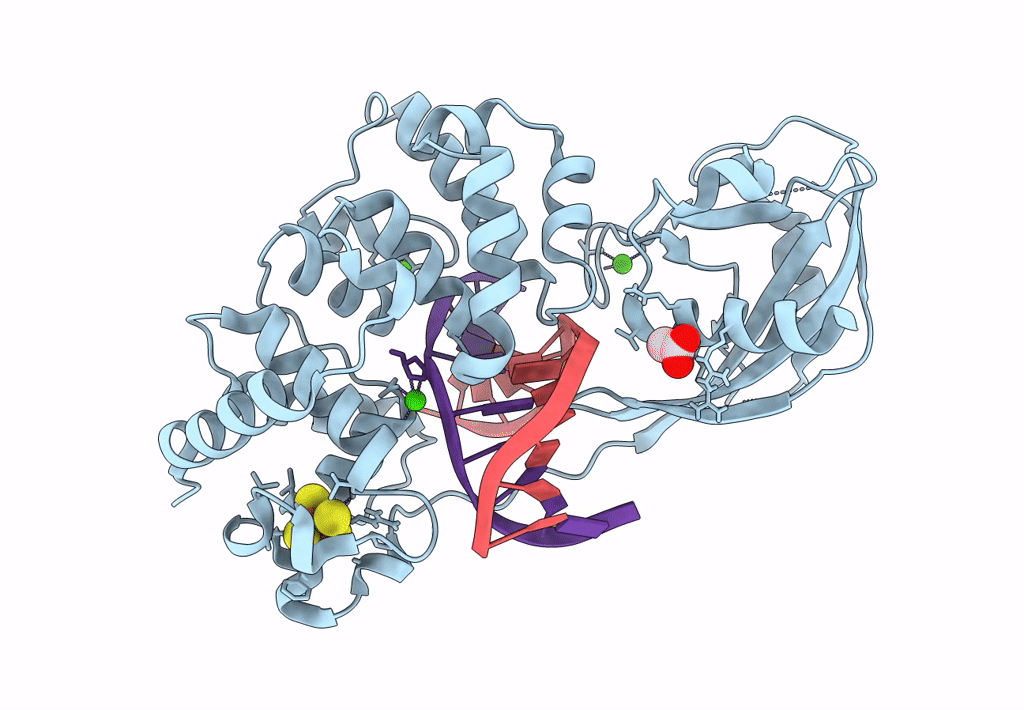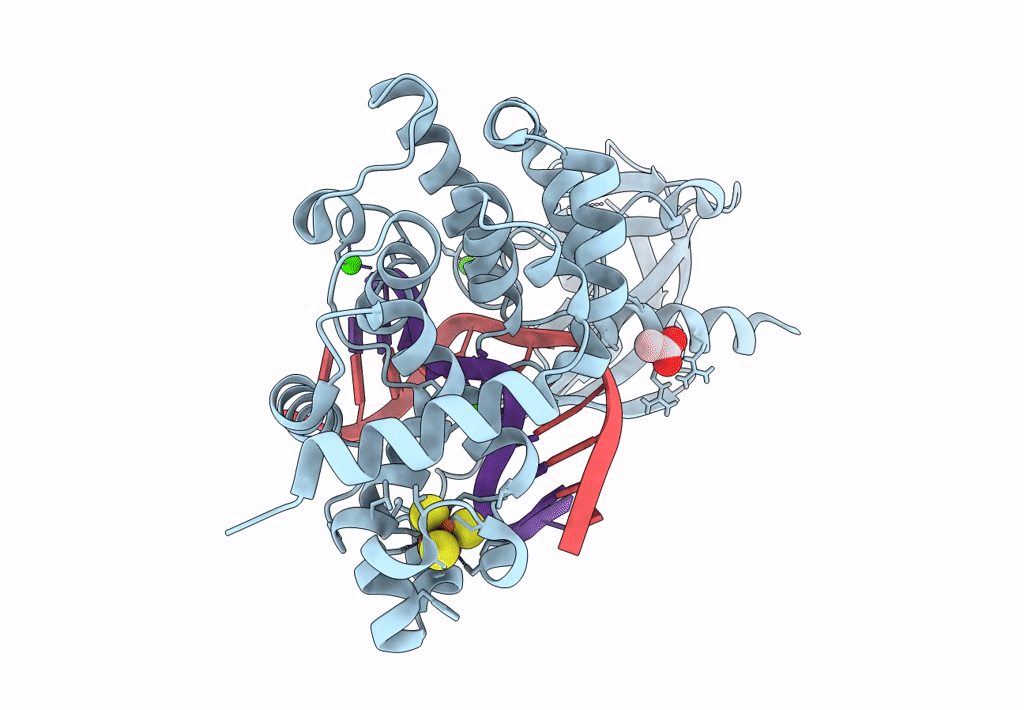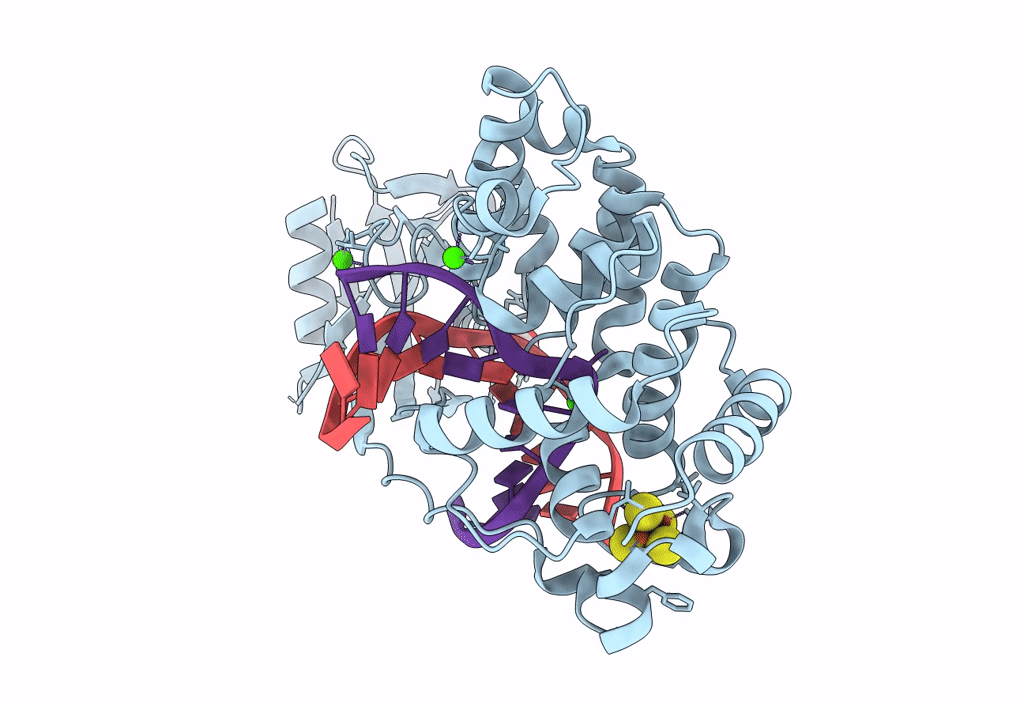
DNA glycosylase MutY variant N146S in complex with DNA containing d(8-oxo-G) paired with an enzyme-generated abasic site product (AP) and crystalized with calcium acetate
Biological Source:
Source Organism(s):
Geobacillus stearothermophilus (Taxon ID: 1422)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.36 Å
R-Value Free:
0.27
R-Value Work:
0.24
R-Value Observed:
0.24
Space Group:
P 21 21 21


