Deposition Date
2022-06-29
Release Date
2023-04-12
Last Version Date
2023-10-25
Entry Detail
PDB ID:
8DJ1
Keywords:
Title:
Crystal structure of NavAb V126T as a basis for the human Nav1.7 Inherited Erythromelalgia S241T mutation
Biological Source:
Source Organism(s):
Aliarcobacter butzleri RM4018 (Taxon ID: 367737)
Expression System(s):
Method Details:
Experimental Method:
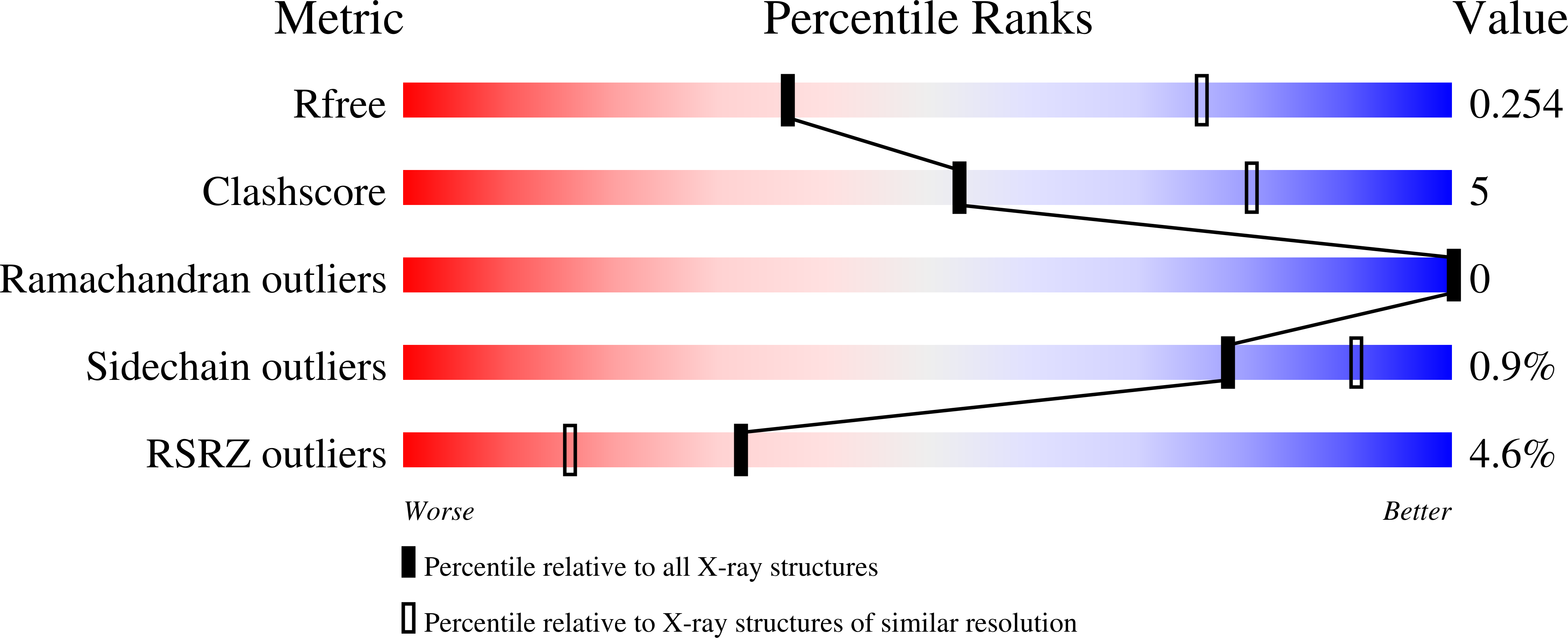
Resolution:
3.10 Å
R-Value Free:
0.25
R-Value Work:
0.21
R-Value Observed:
0.21
Space Group:
I 4 2 2