Deposition Date
2022-06-21
Release Date
2022-08-31
Last Version Date
2024-10-23
Entry Detail
PDB ID:
8DF7
Keywords:
Title:
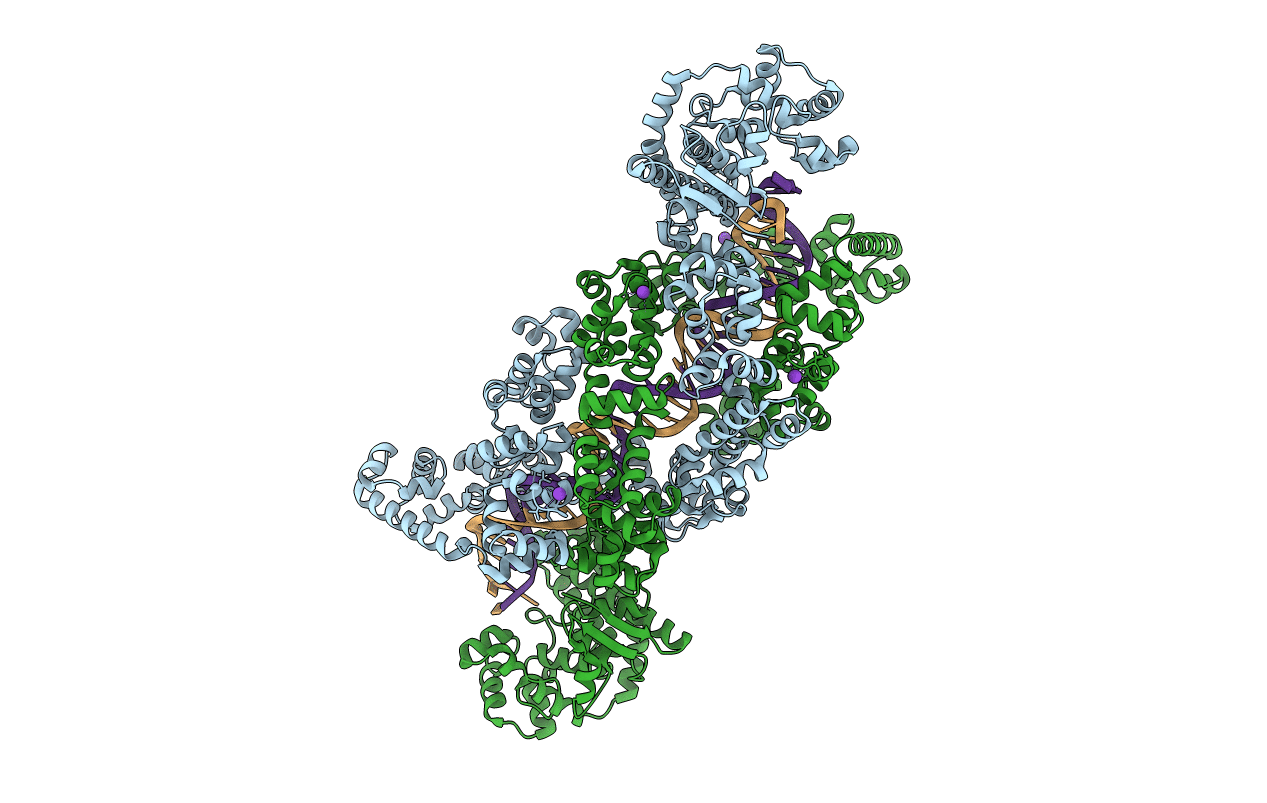
Structure of M. kandleri topoisomerase V in complex with DNA. 38 base pair symmetric DNA complex
Biological Source:
Source Organism(s):
Methanopyrus kandleri (Taxon ID: 2320)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
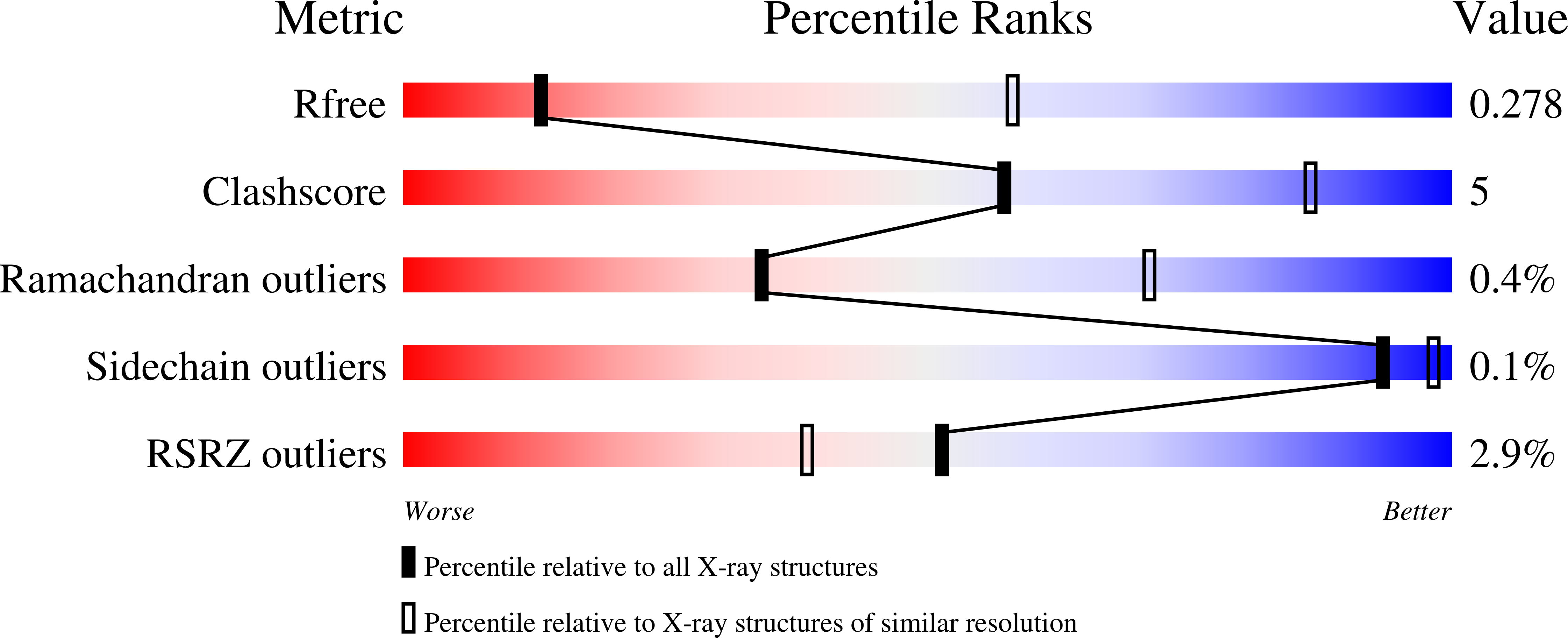
Resolution:
3.52 Å
R-Value Free:
0.27
R-Value Work:
0.22
R-Value Observed:
0.23
Space Group:
P 43 21 2