Deposition Date
2022-06-18
Release Date
2022-08-31
Last Version Date
2023-10-25
Entry Detail
PDB ID:
8DDI
Keywords:
Title:
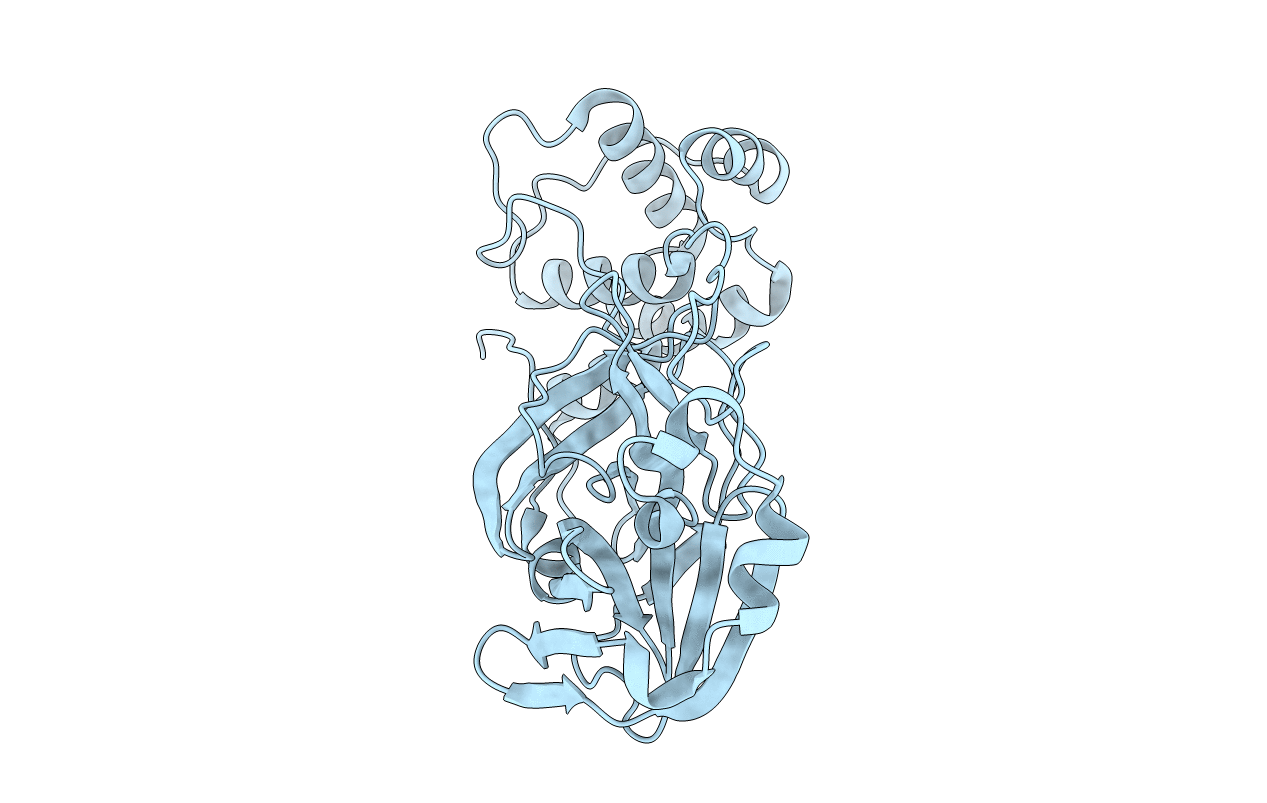
Crystal Structure of SARS-CoV-2 Main Protease (Mpro) E166N Mutant
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
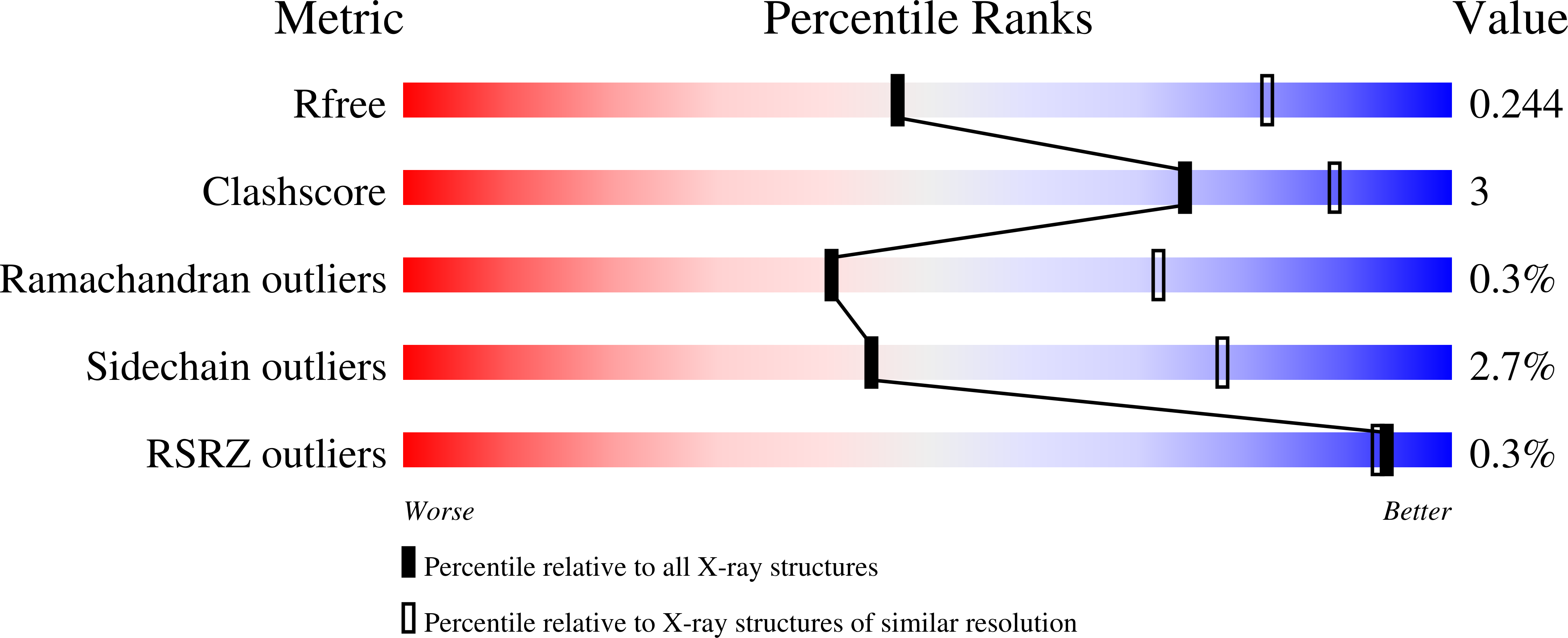
Resolution:
2.80 Å
R-Value Free:
0.24
R-Value Work:
0.17
R-Value Observed:
0.18
Space Group:
C 1 2 1