Deposition Date
2023-01-21
Release Date
2023-08-16
Last Version Date
2024-03-27
Entry Detail
PDB ID:
8C8V
Keywords:
Title:
Priestia megaterium mupirocin-resistant isoleucyl-tRNA synthetase 2 with a fully-resolved C-terminal tRNA-binding domain complexed with an isoleucyl-adenylate analogue
Biological Source:
Source Organism(s):
Priestia megaterium NBRC 15308 = ATCC 14581 (Taxon ID: 1348623)
Expression System(s):
Method Details:
Experimental Method:
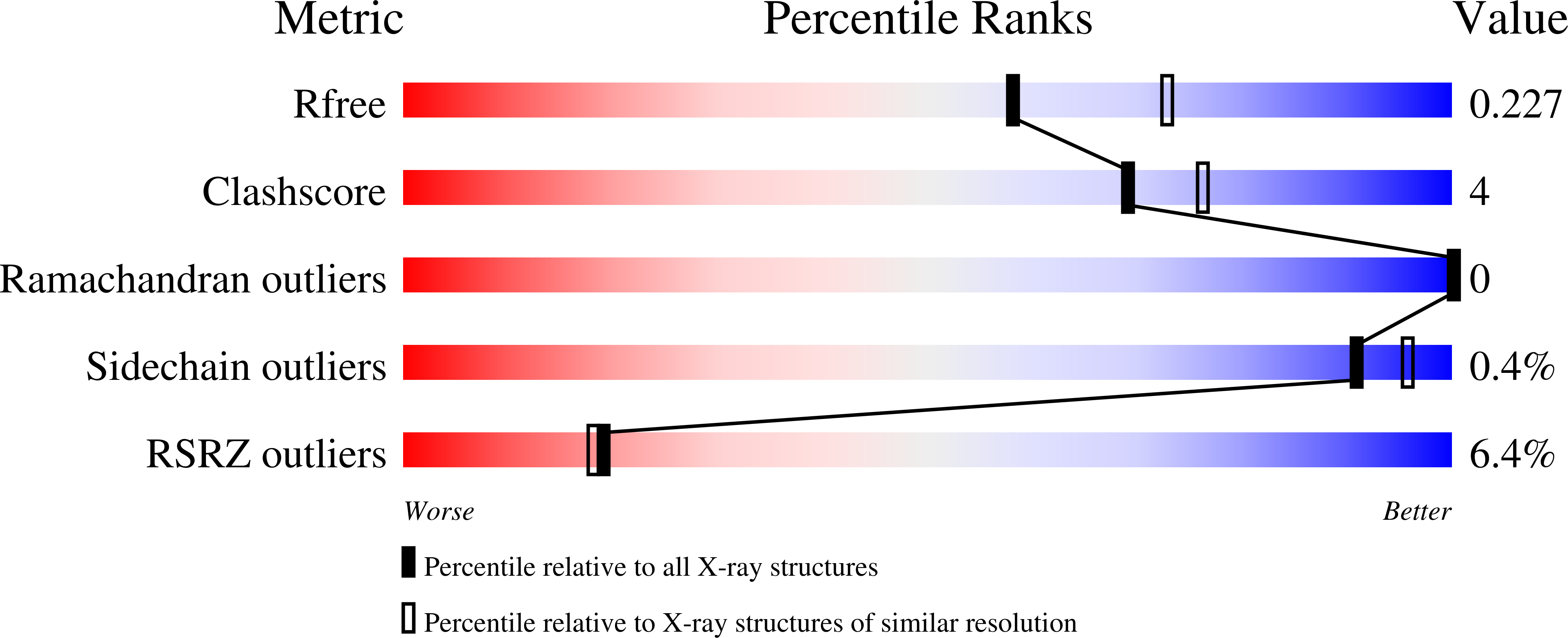
Resolution:
2.20 Å
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 43 21 2