Deposition Date
2023-01-11
Release Date
2024-01-31
Last Version Date
2024-02-28
Entry Detail
PDB ID:
8C64
Keywords:
Title:
Crystal structure of arsenoplatin-1/B-DNA adduct obtained upon 48 h of soaking
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Resolution:
2.51 Å
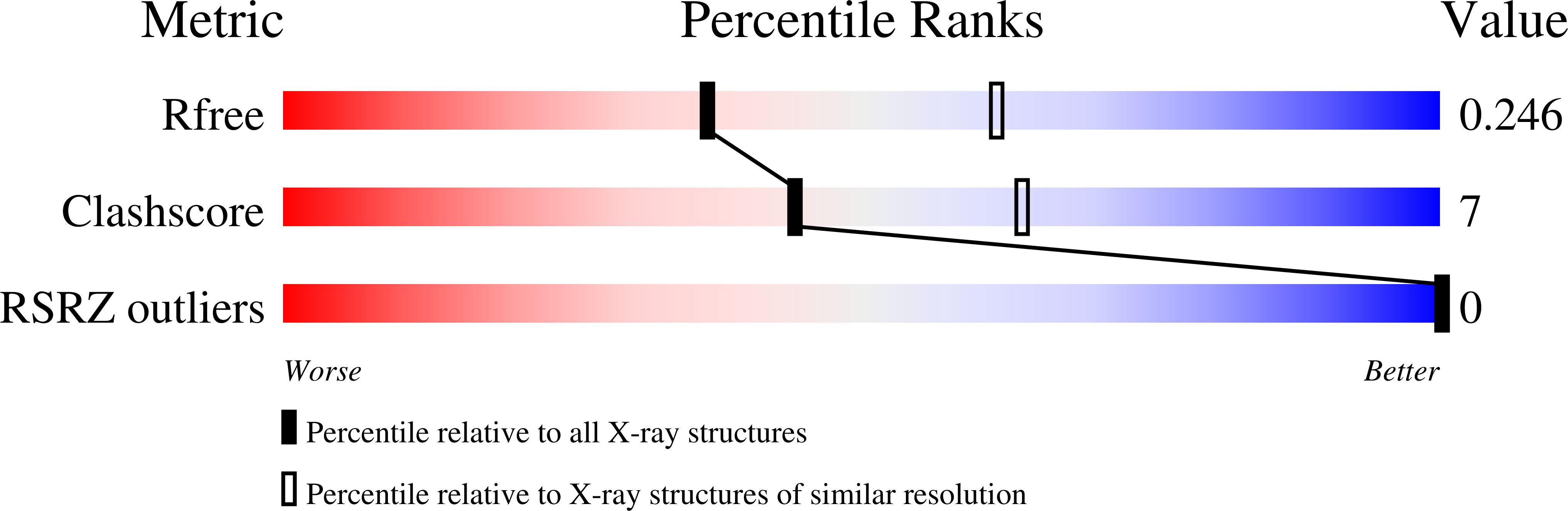
R-Value Free:
0.23
R-Value Work:
0.17
R-Value Observed:
0.18
Space Group:
P 21 21 21