Deposition Date
2022-09-27
Release Date
2022-11-30
Last Version Date
2024-06-19
Entry Detail
PDB ID:
8B6X
Keywords:
Title:
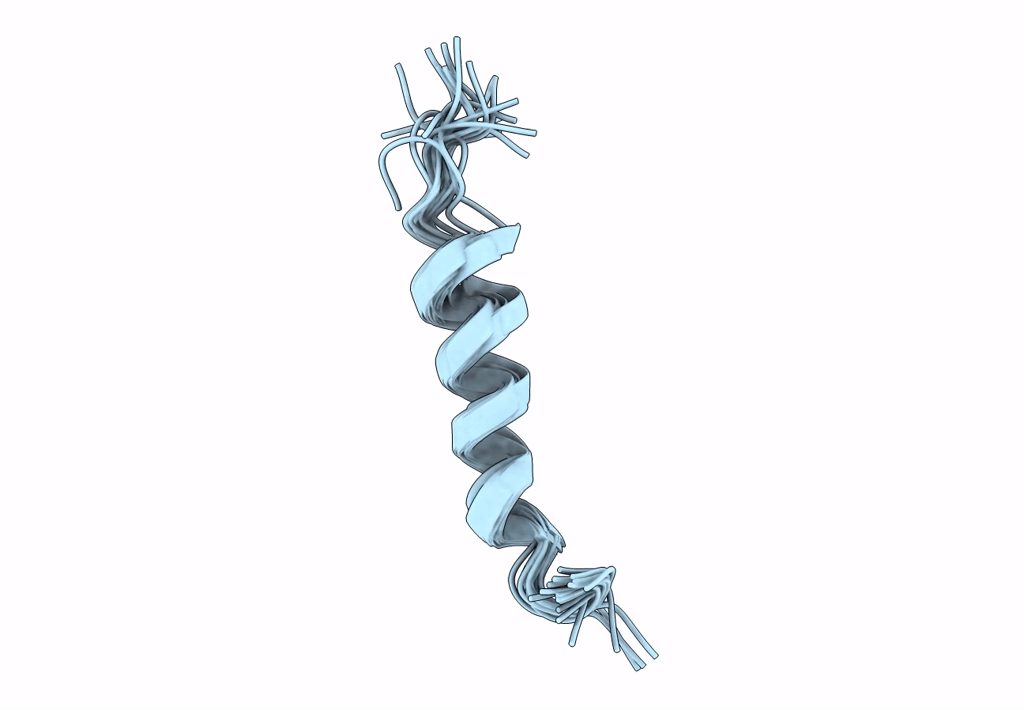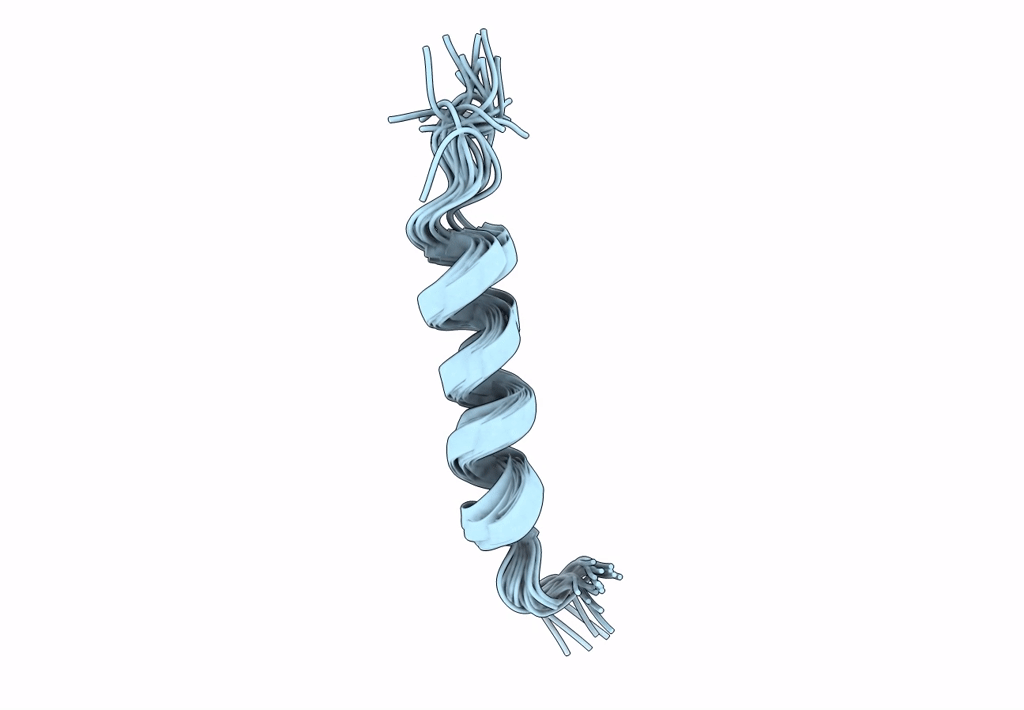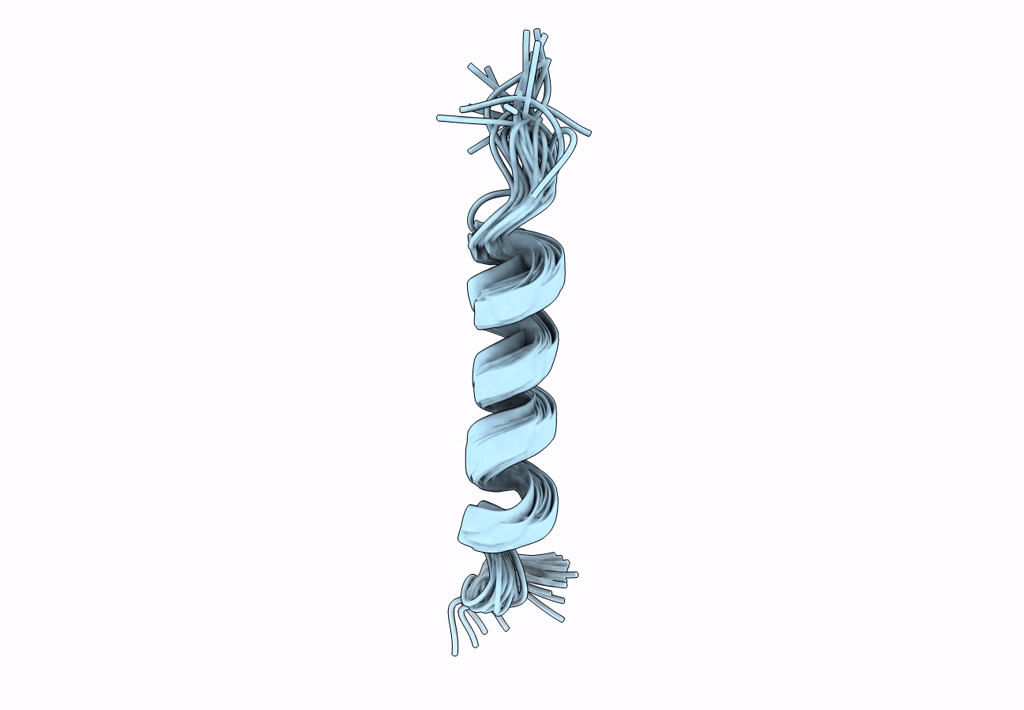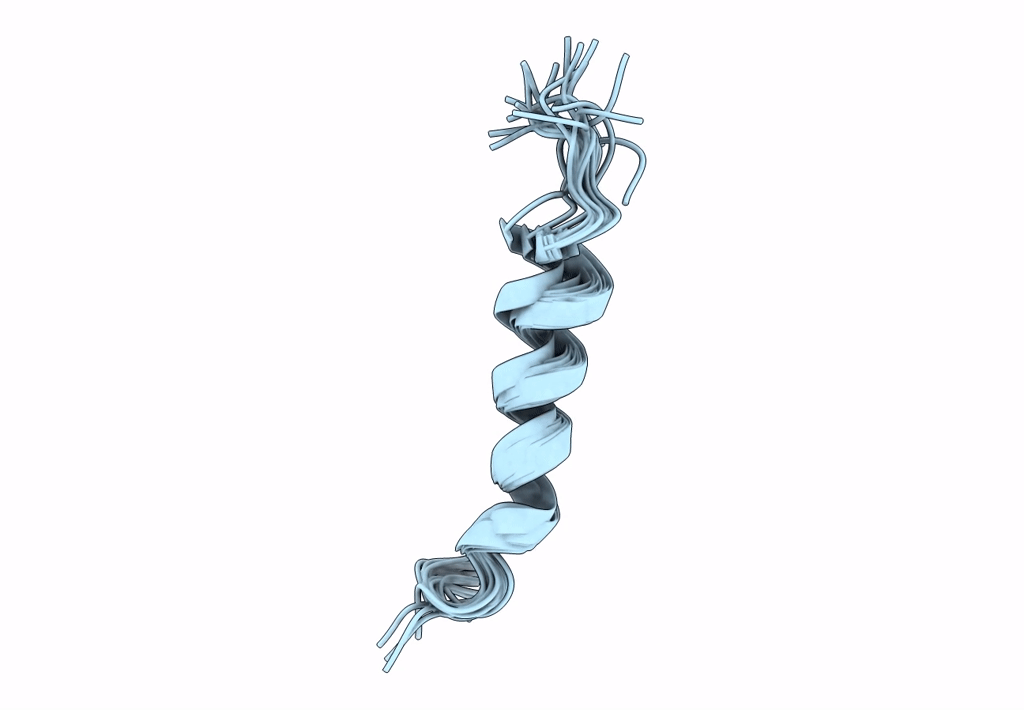
NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in DPC micelles
Biological Source:
Source Organism(s):
Human immunodeficiency virus 1 (Taxon ID: 11676)
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
target function


