Deposition Date
2022-06-19
Release Date
2023-02-15
Last Version Date
2025-12-17
Entry Detail
PDB ID:
8A6T
Keywords:
Title:
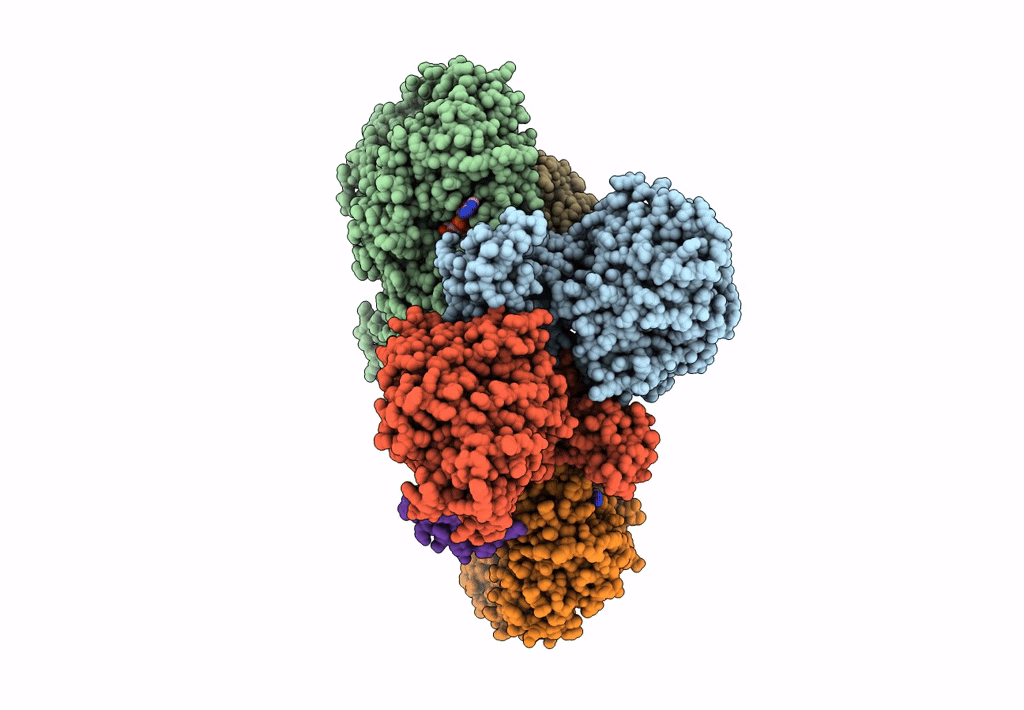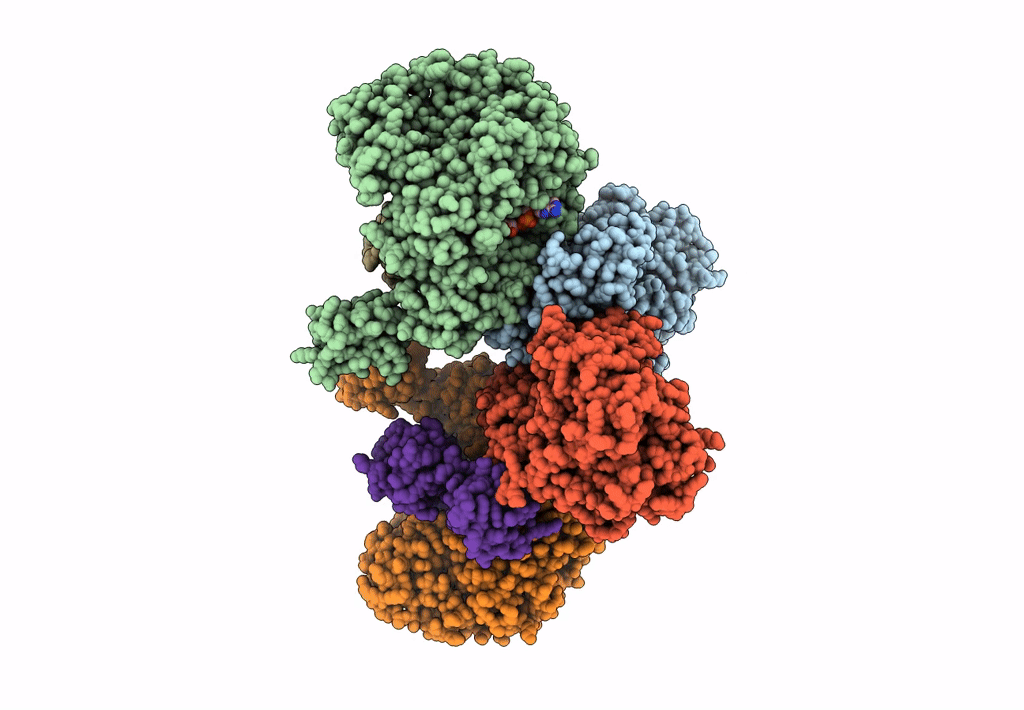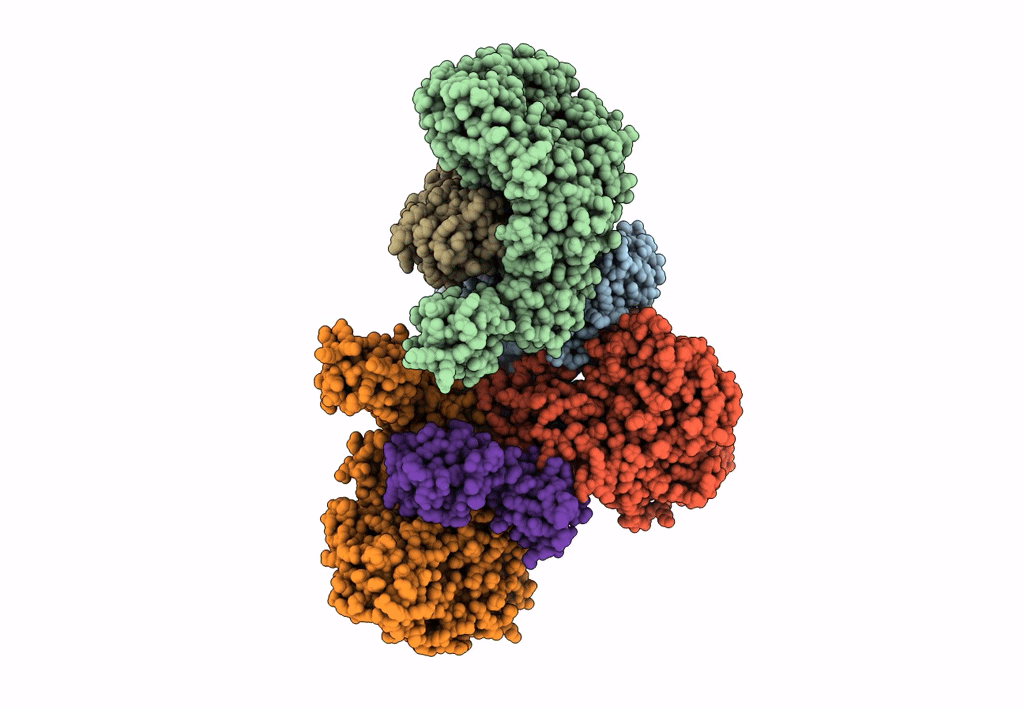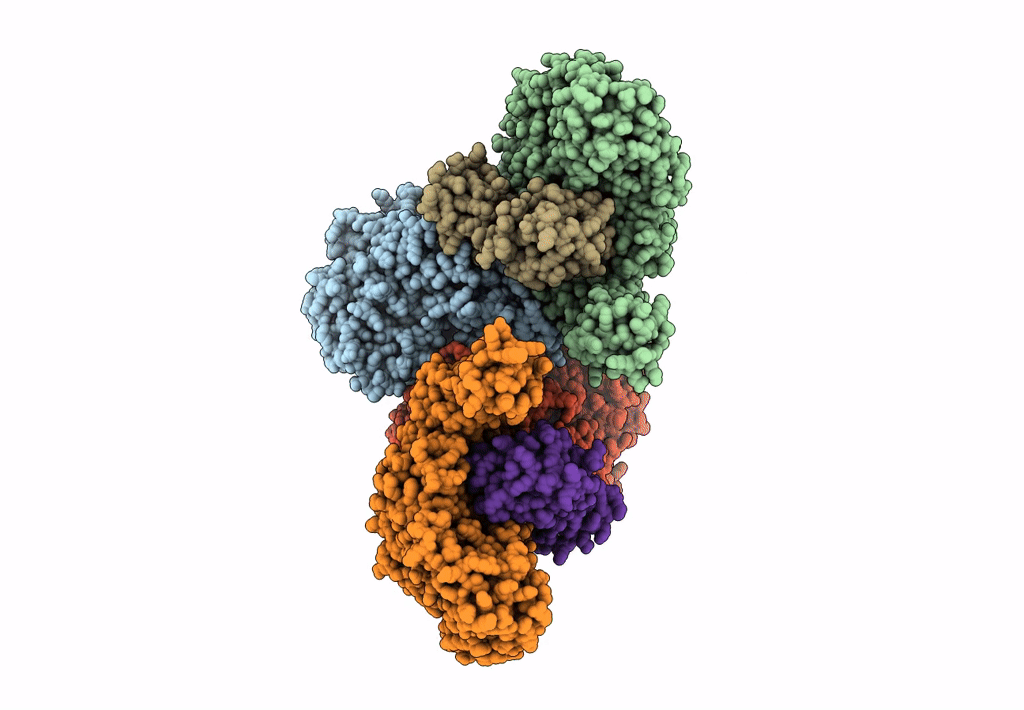
Cryo-EM structure of the electron bifurcating Fe-Fe hydrogenase HydABC complex from Thermoanaerobacter kivui in the reduced state
Biological Source:
Source Organism:
Thermoanaerobacter kivui (Taxon ID: 2325)
Method Details:
Experimental Method:
Resolution:
3.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


