Deposition Date
2022-06-15
Release Date
2022-12-14
Last Version Date
2024-07-24
Entry Detail
PDB ID:
8A5O
Keywords:
Title:
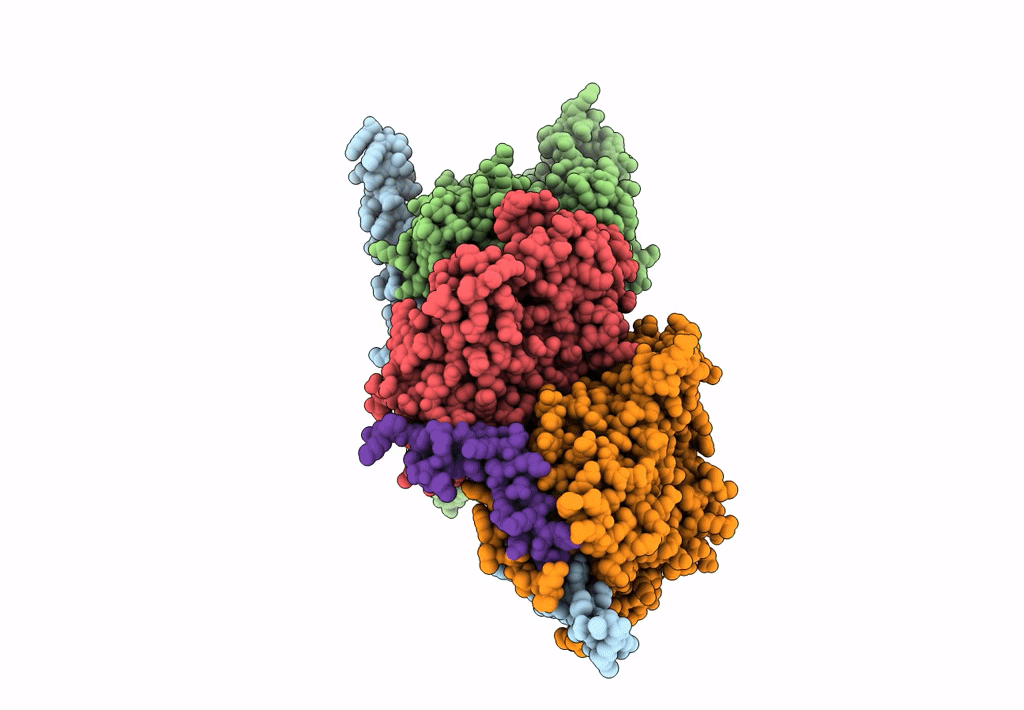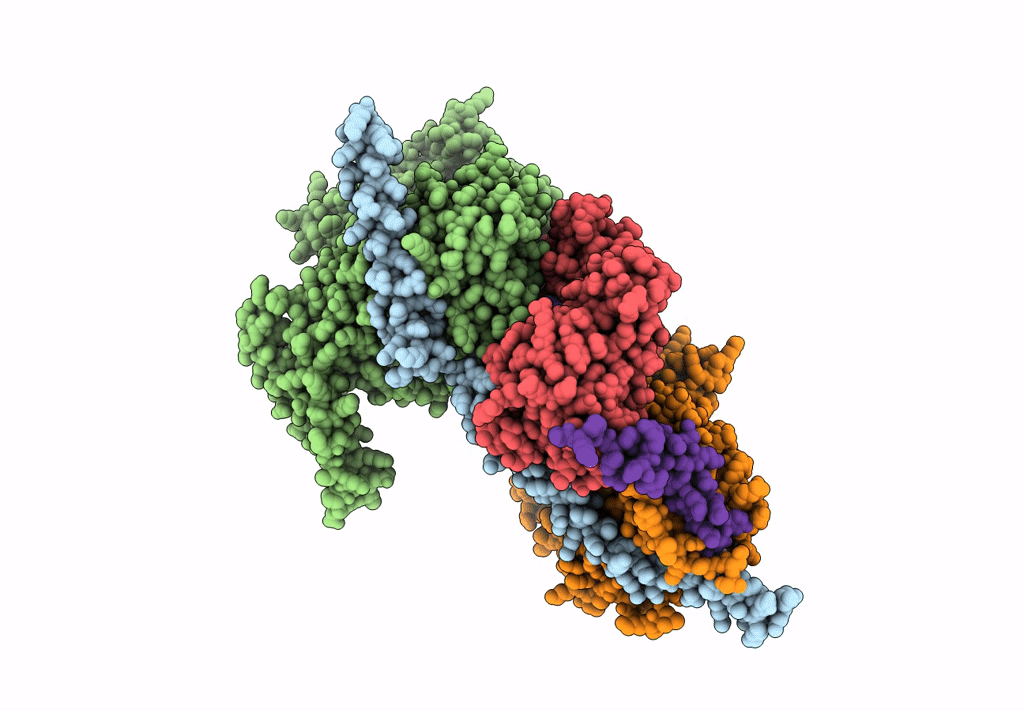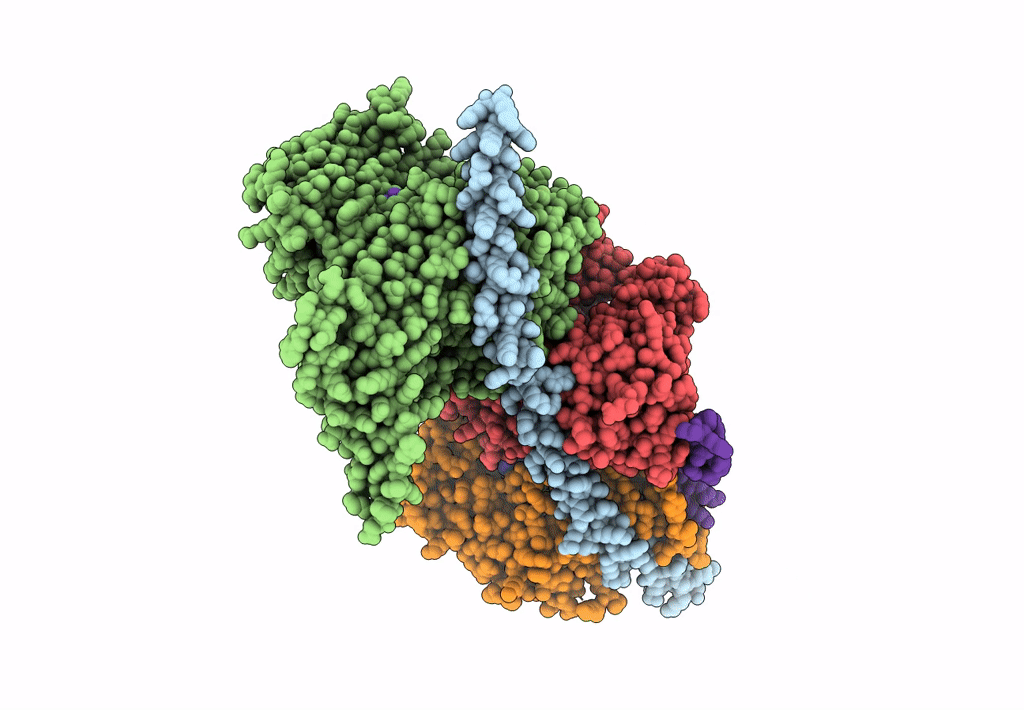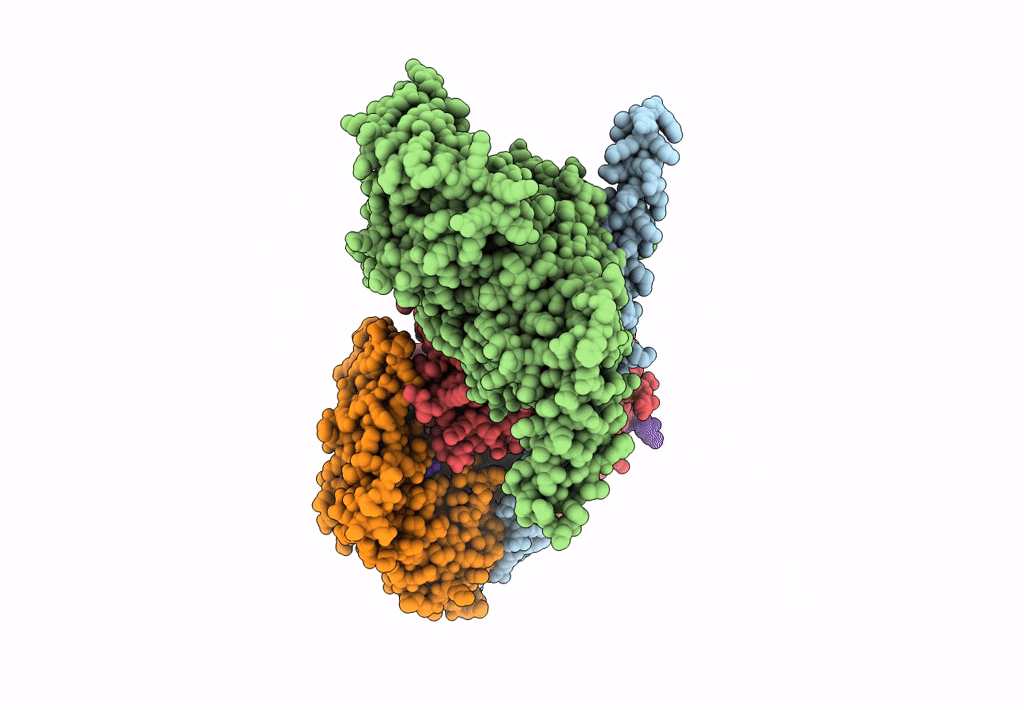
Structure of Arp4-Ies4-N-actin-Arp8-Ino80HSA subcomplex (A-module) of S. cerevisiae INO80
Biological Source:
Source Organism:
Saccharomyces cerevisiae S288C (Taxon ID: 559292)
Host Organism:
Method Details:
Experimental Method:
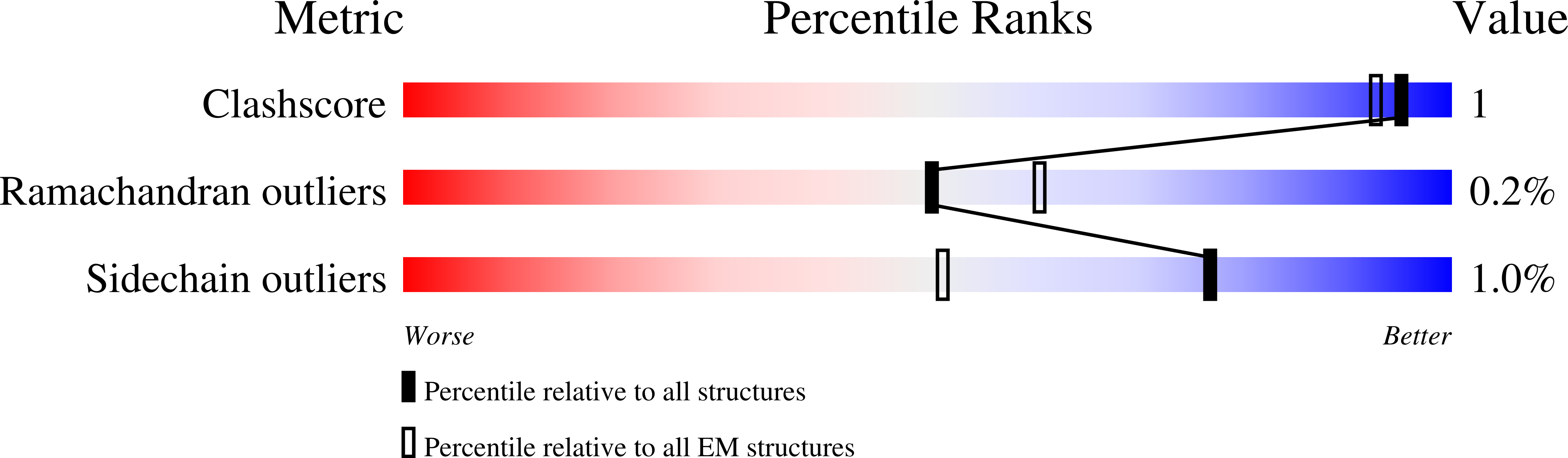
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE