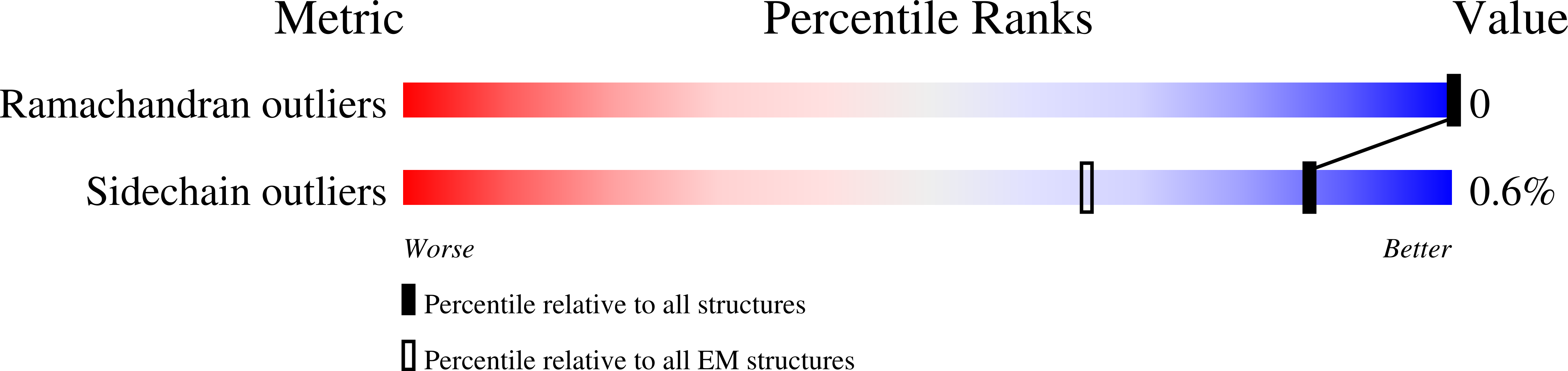
Deposition Date
2022-04-19
Release Date
2022-11-30
Last Version Date
2024-11-13
Entry Detail
PDB ID:
7ZME
Keywords:
Title:
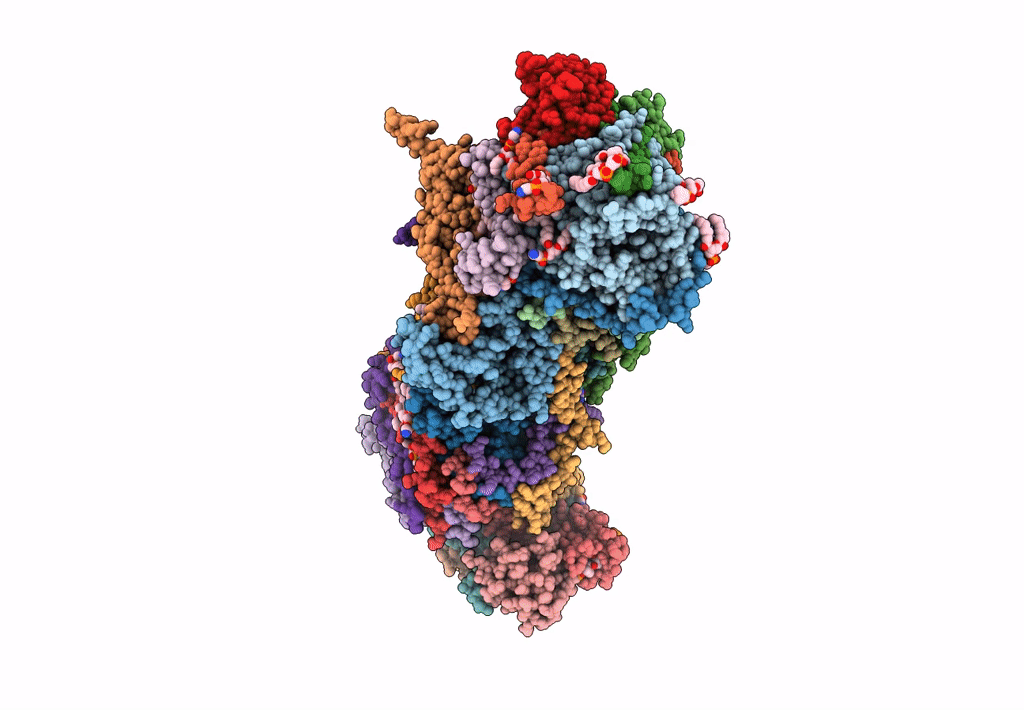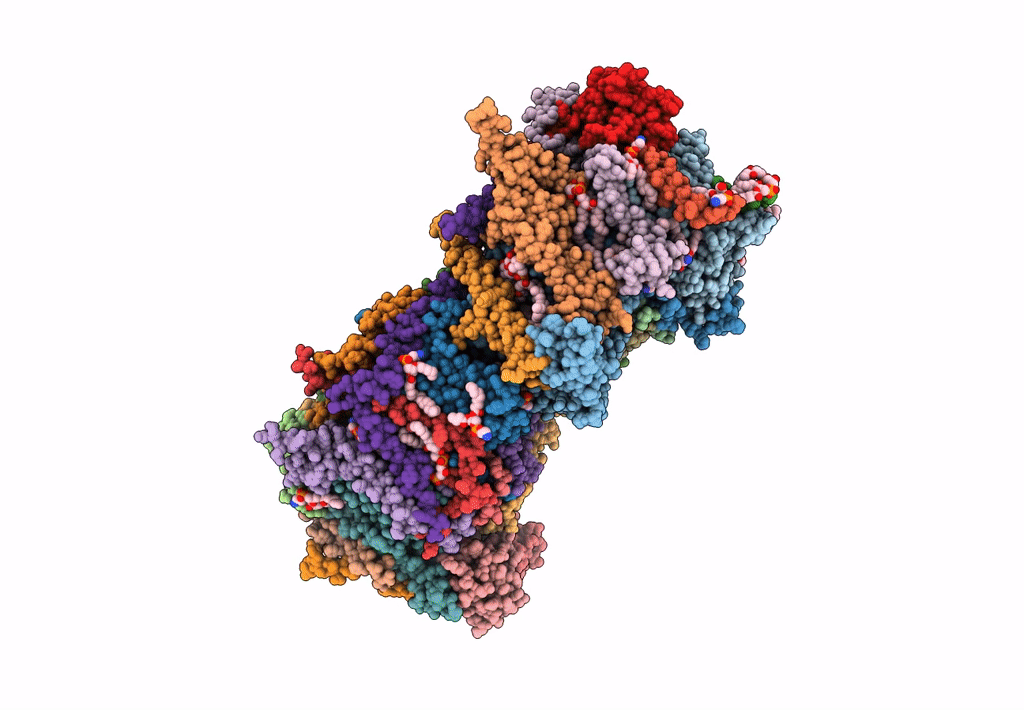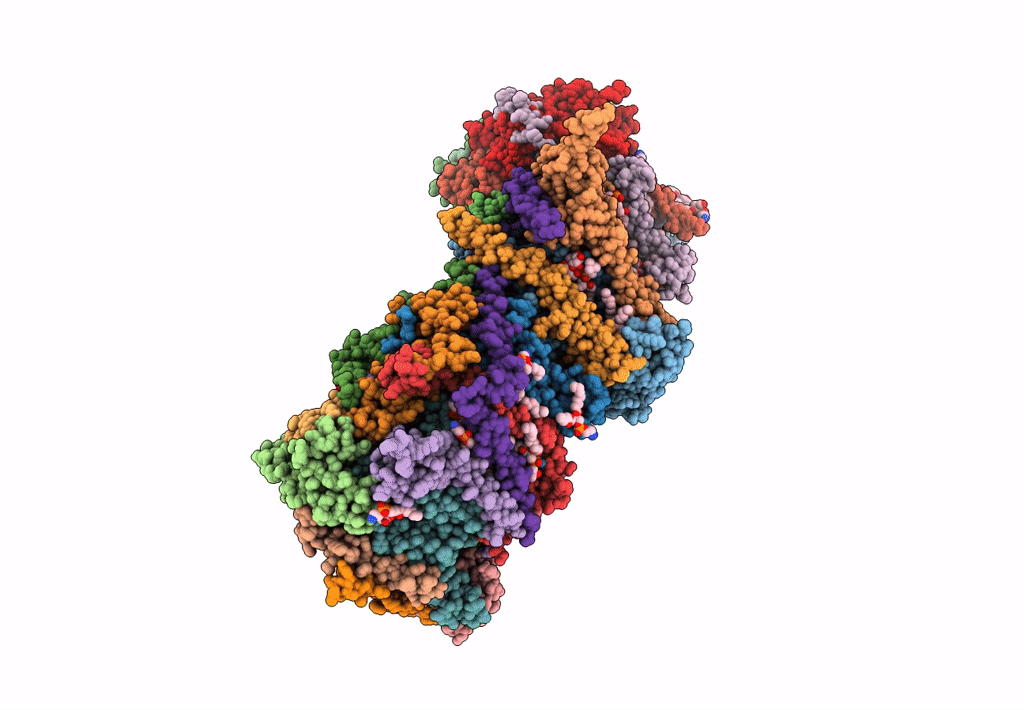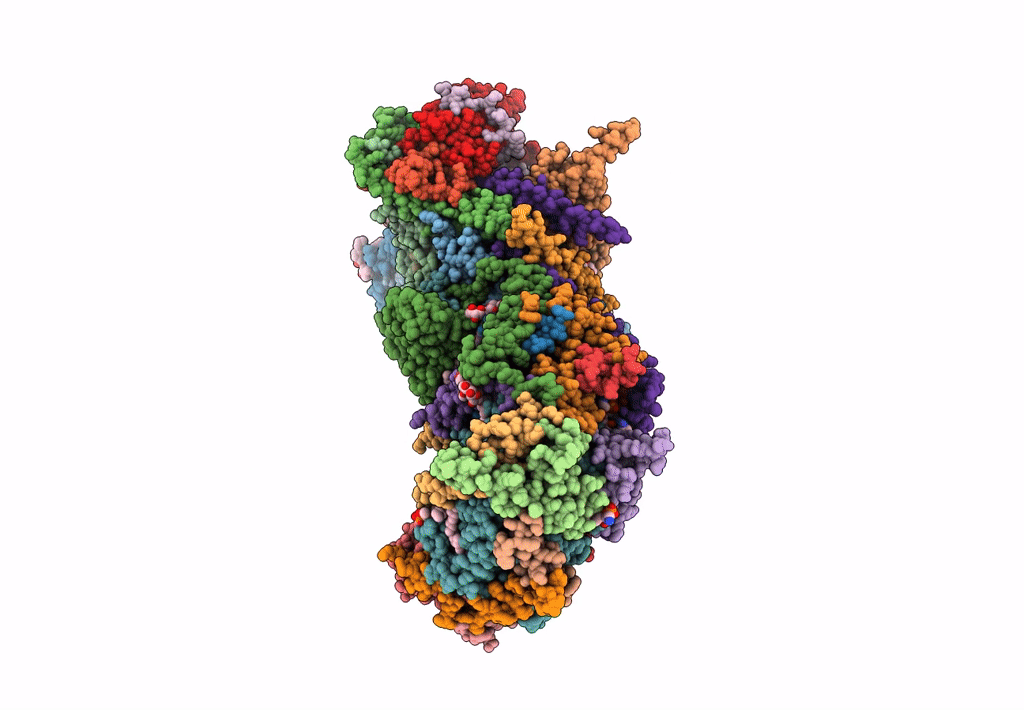
CryoEM structure of mitochondrial complex I from Chaetomium thermophilum (state 2) - membrane arm
Biological Source:
Source Organism(s):
Method Details:
Experimental Method:
Resolution:
2.83 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE