Deposition Date
2022-03-22
Release Date
2022-07-13
Last Version Date
2024-11-13
Entry Detail
PDB ID:
7ZAI
Keywords:
Title:
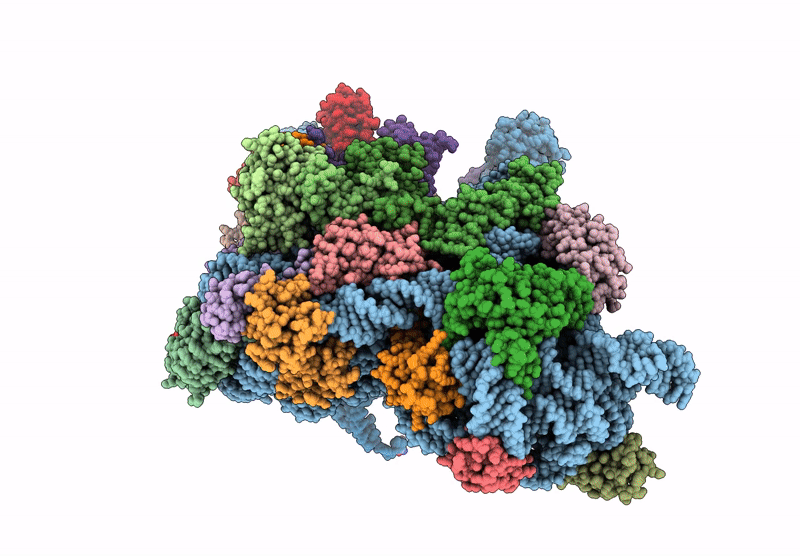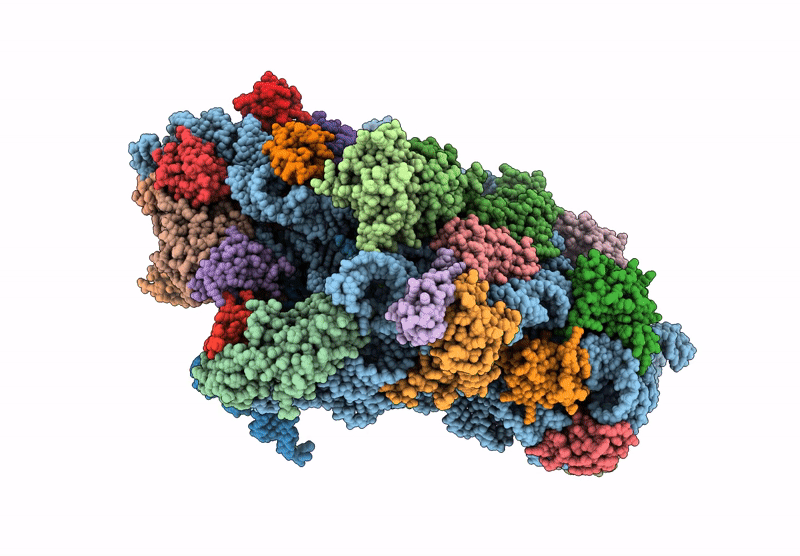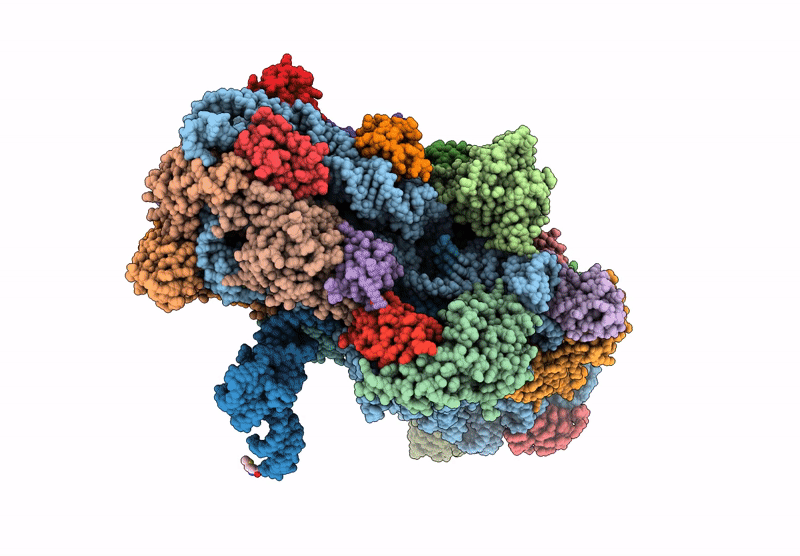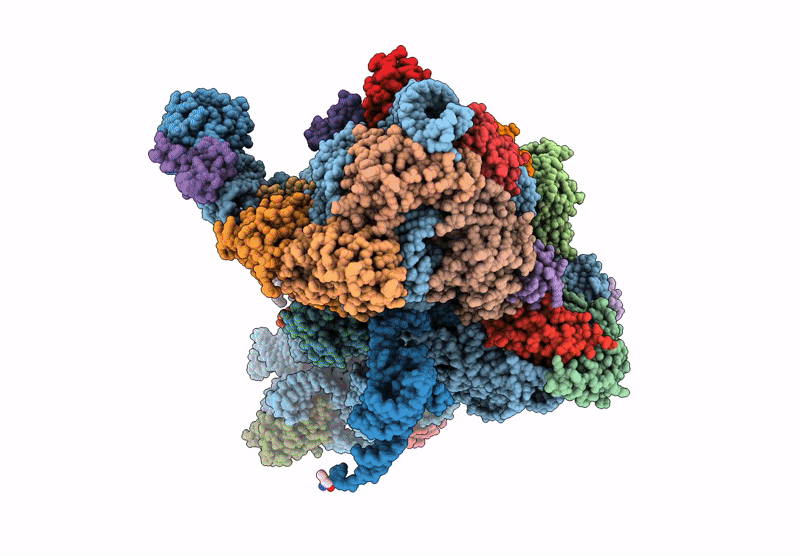
Cryo-EM structure of a Pyrococcus abyssi 30S bound to Met-initiator tRNA, mRNA and aIF1A.
Biological Source:
Source Organism(s):
Pyrococcus abyssi GE5 (Taxon ID: 272844)
Pyrococcus abyssi (Taxon ID: 29292)
Pyrococcus abyssi (Taxon ID: 29292)
Expression System(s):
Method Details:
Experimental Method:
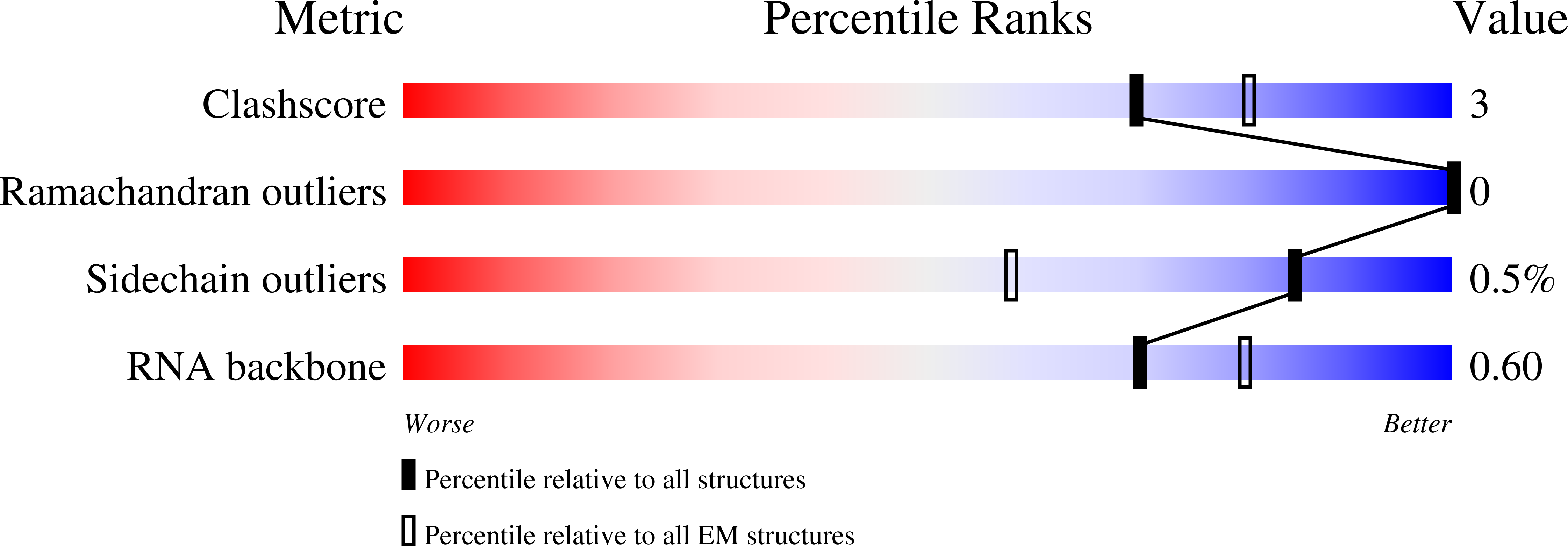
Resolution:
2.60 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE