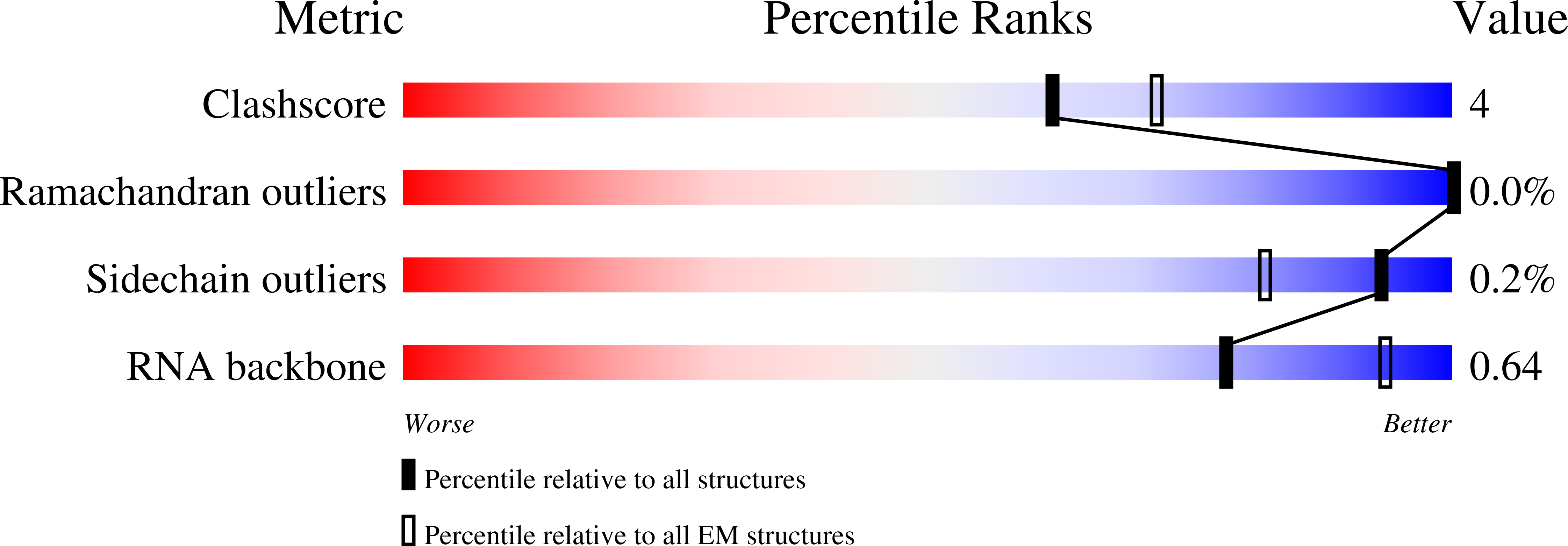
Deposition Date
2022-02-24
Release Date
2022-08-31
Last Version Date
2025-10-01
Entry Detail
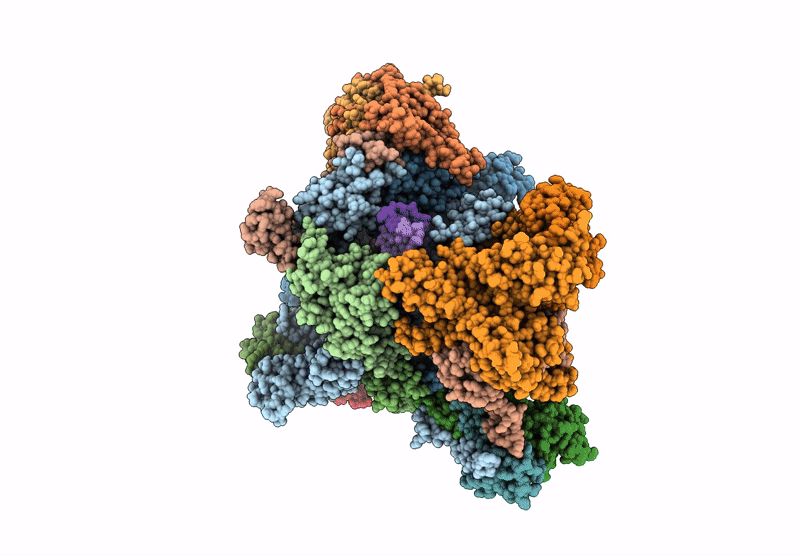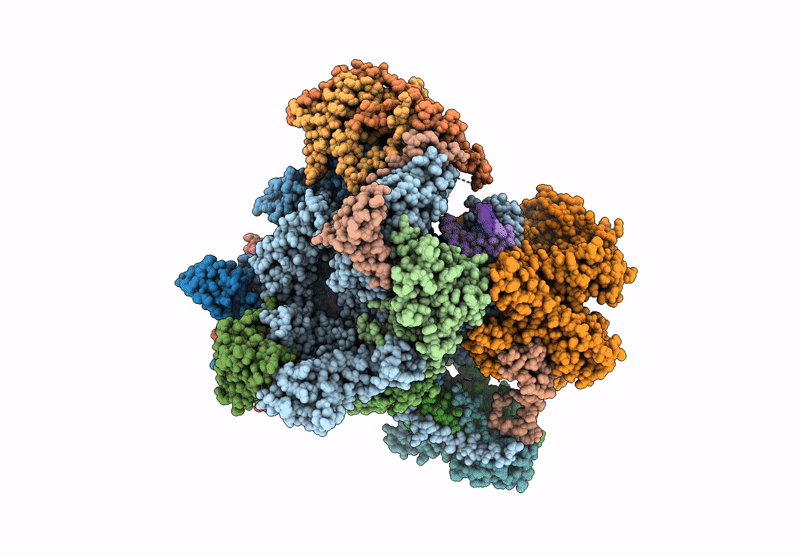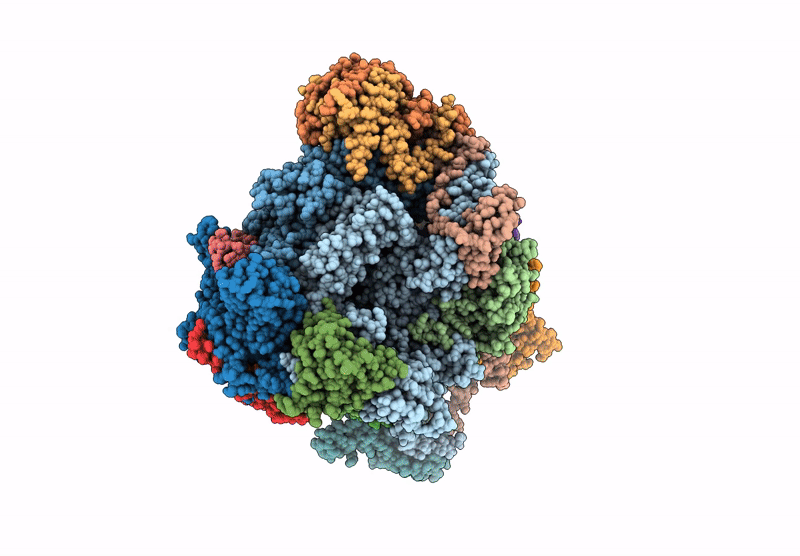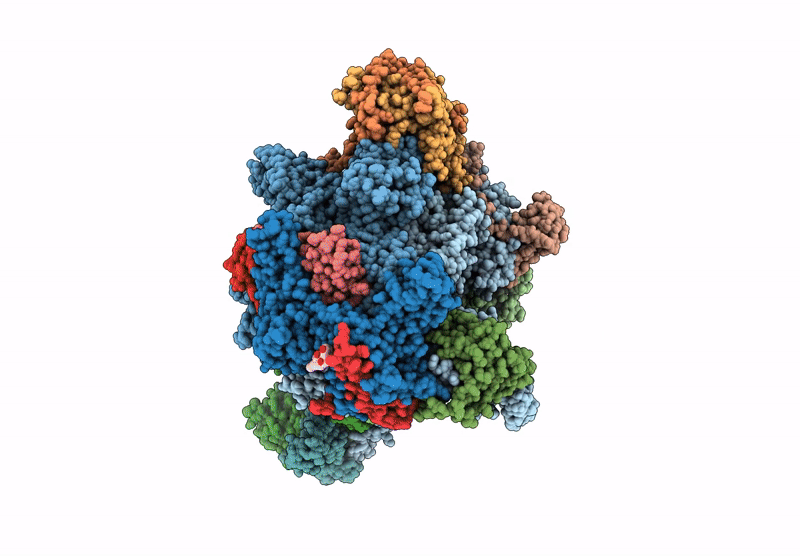
PDB ID:
7Z1M
Keywords:
Title:
Structure of yeast RNA Polymerase III Elongation Complex (EC)
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Saccharomyces cerevisiae W303 (Taxon ID: 580240)
Saccharomyces cerevisiae W303 (Taxon ID: 580240)
Method Details:
Experimental Method:
Resolution:
3.40 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE