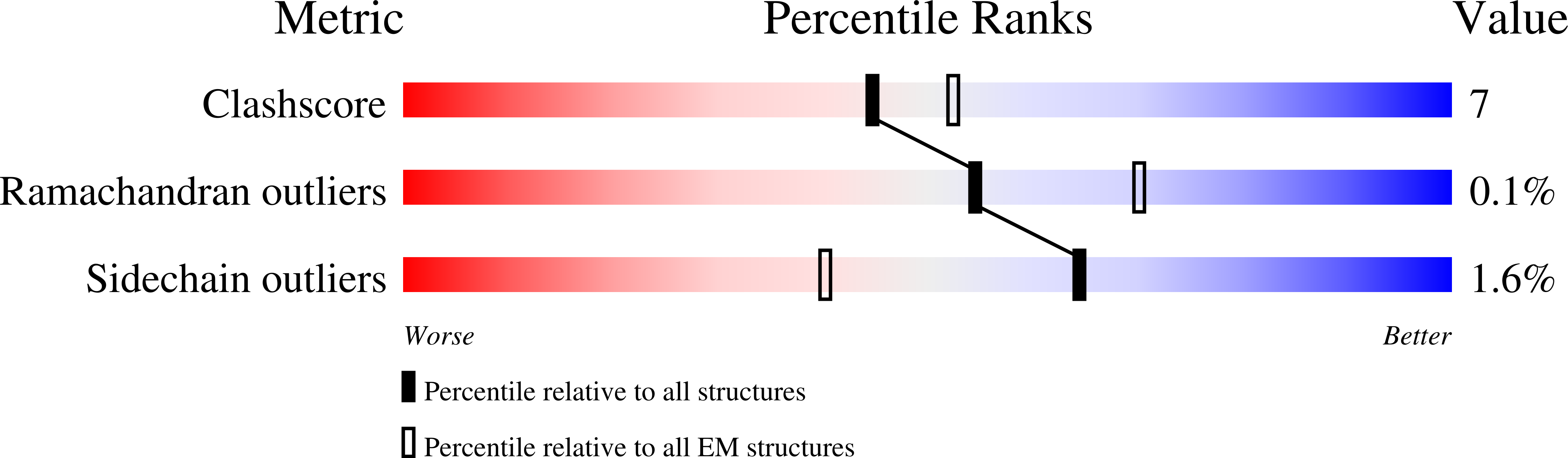
Deposition Date
2022-02-23
Release Date
2022-09-28
Last Version Date
2024-07-17
Entry Detail
PDB ID:
7Z0S
Keywords:
Title:
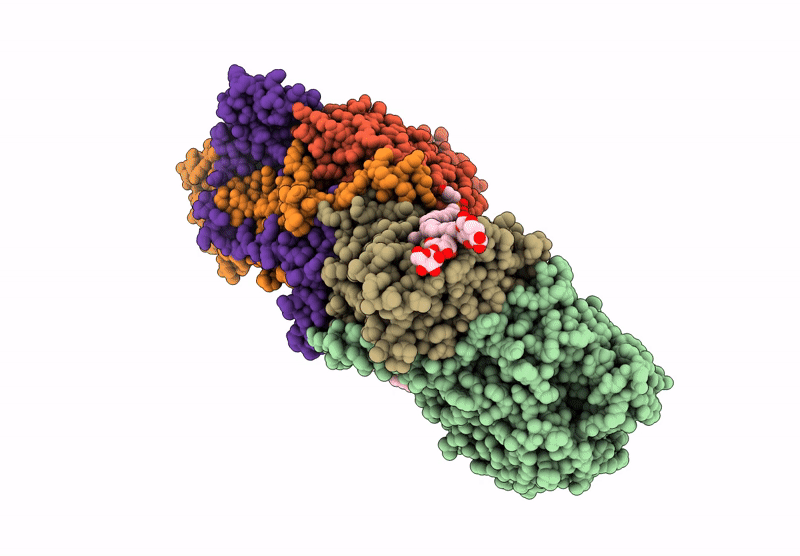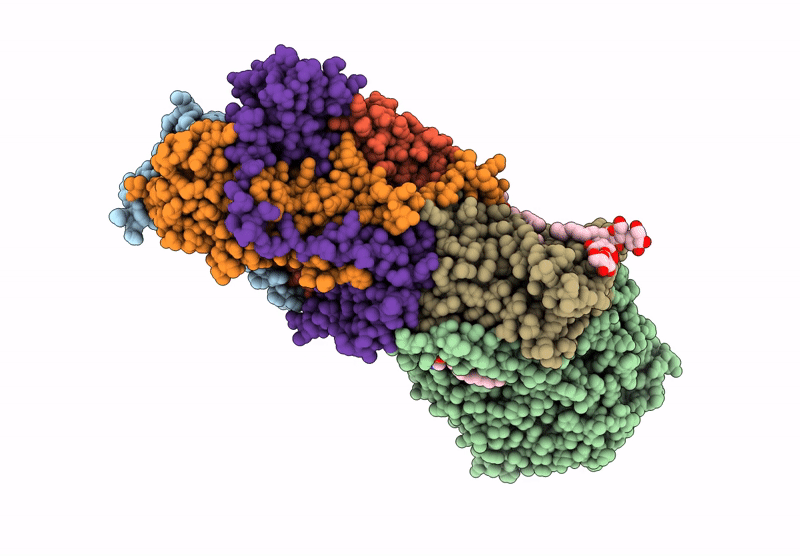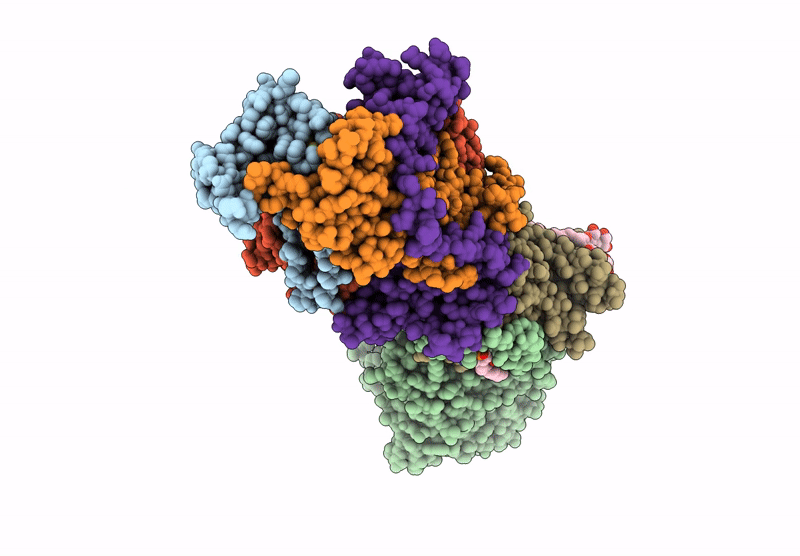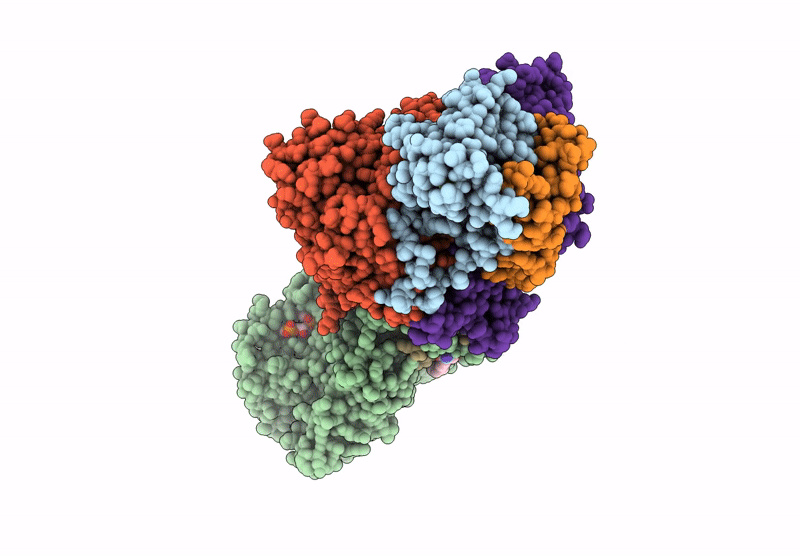
Structure of the Escherichia coli formate hydrogenlyase complex (anaerobic preparation, without formate dehydrogenase H)
Biological Source:
Source Organism(s):
Escherichia coli K-12 (Taxon ID: 83333)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.60 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE