Deposition Date
2022-02-22
Release Date
2022-11-23
Last Version Date
2024-11-06
Entry Detail
PDB ID:
7Z04
Keywords:
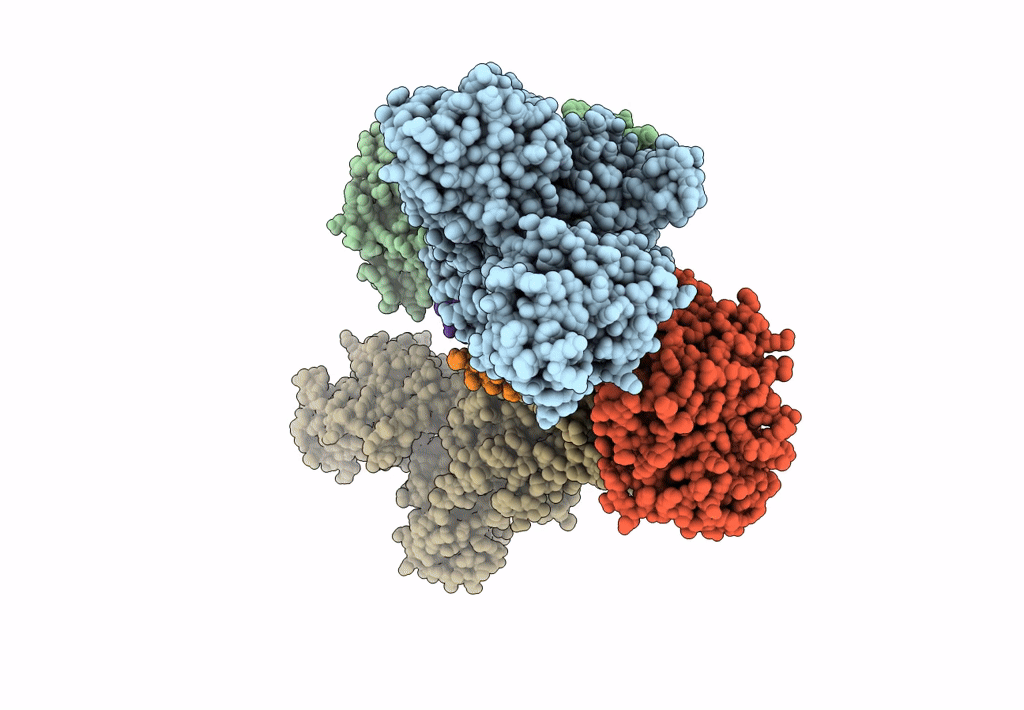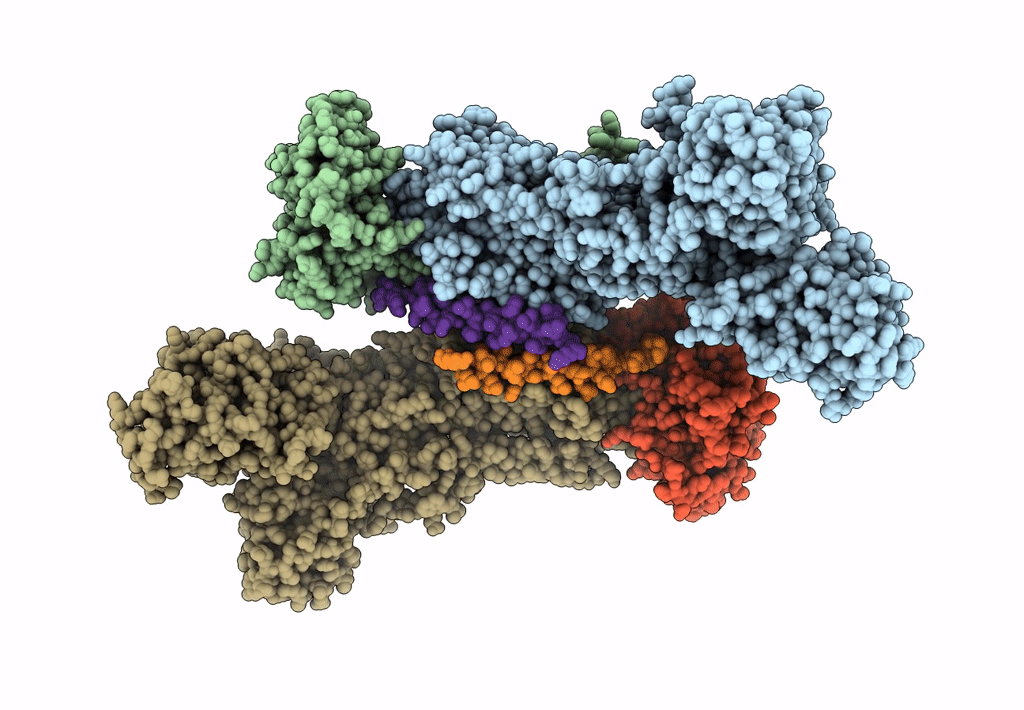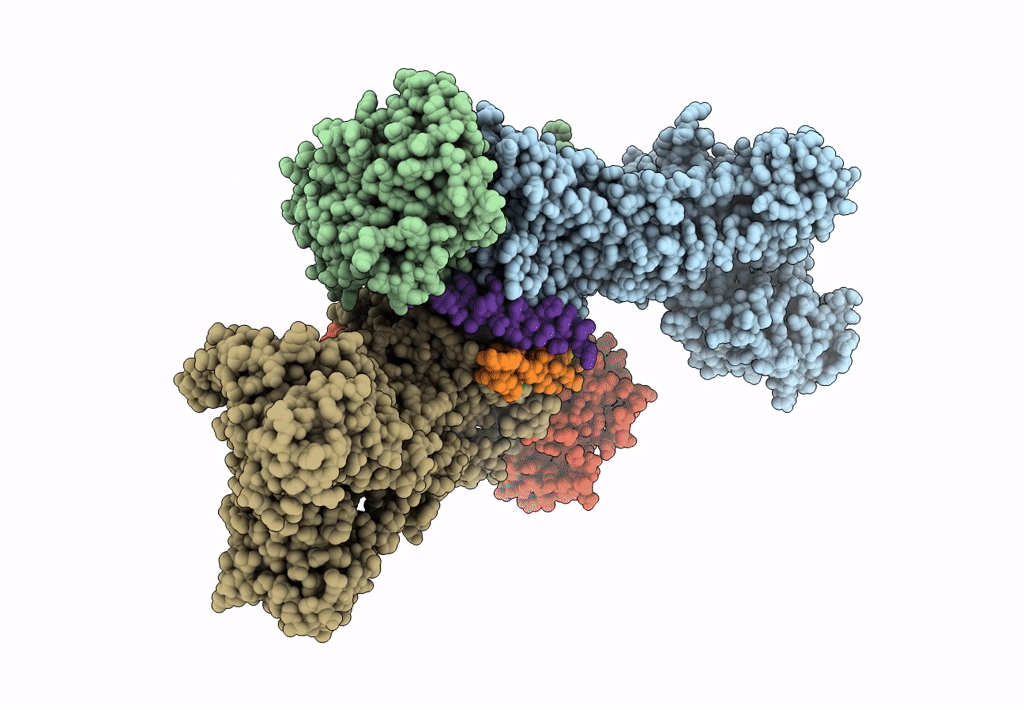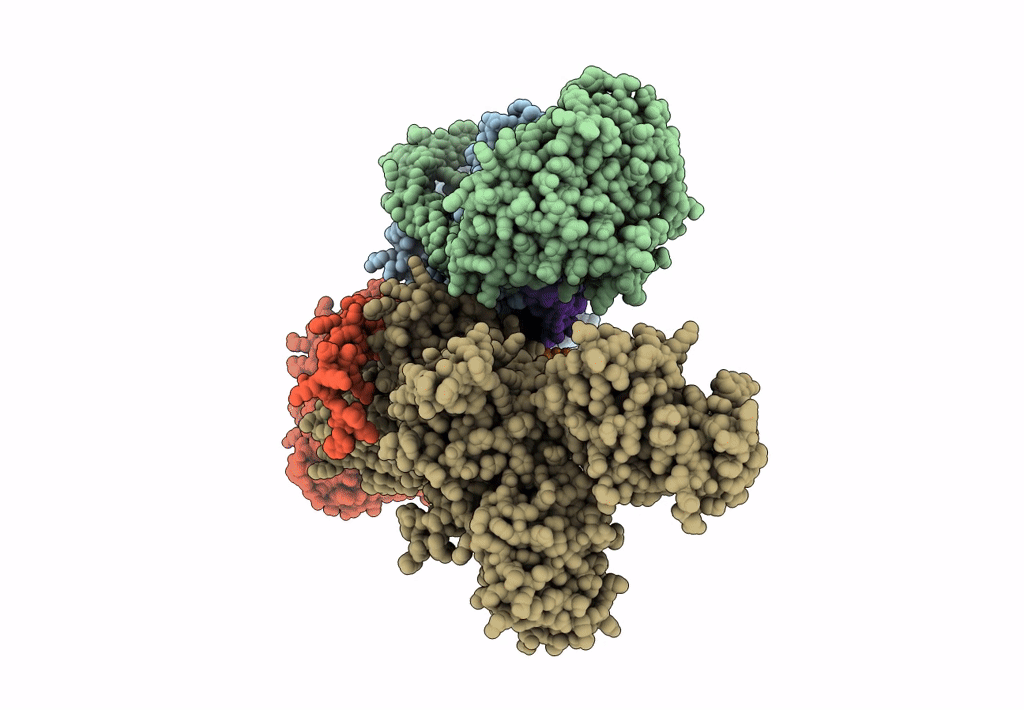
Title:
10 mM Rb+ soak of beryllium fluoride inhibited Na+,K+-ATPase, E2-BeFx (rigid body model)
Biological Source:
Source Organism(s):
Sus scrofa (Taxon ID: 9823)
Method Details:
Experimental Method:
Resolution:
7.50 Å
R-Value Free:
0.33
R-Value Work:
0.30
R-Value Observed:
0.30
Space Group:
P 21 21 21


