Deposition Date
2022-06-24
Release Date
2022-08-17
Last Version Date
2024-11-06
Entry Detail
PDB ID:
7Y96
Keywords:
Title:
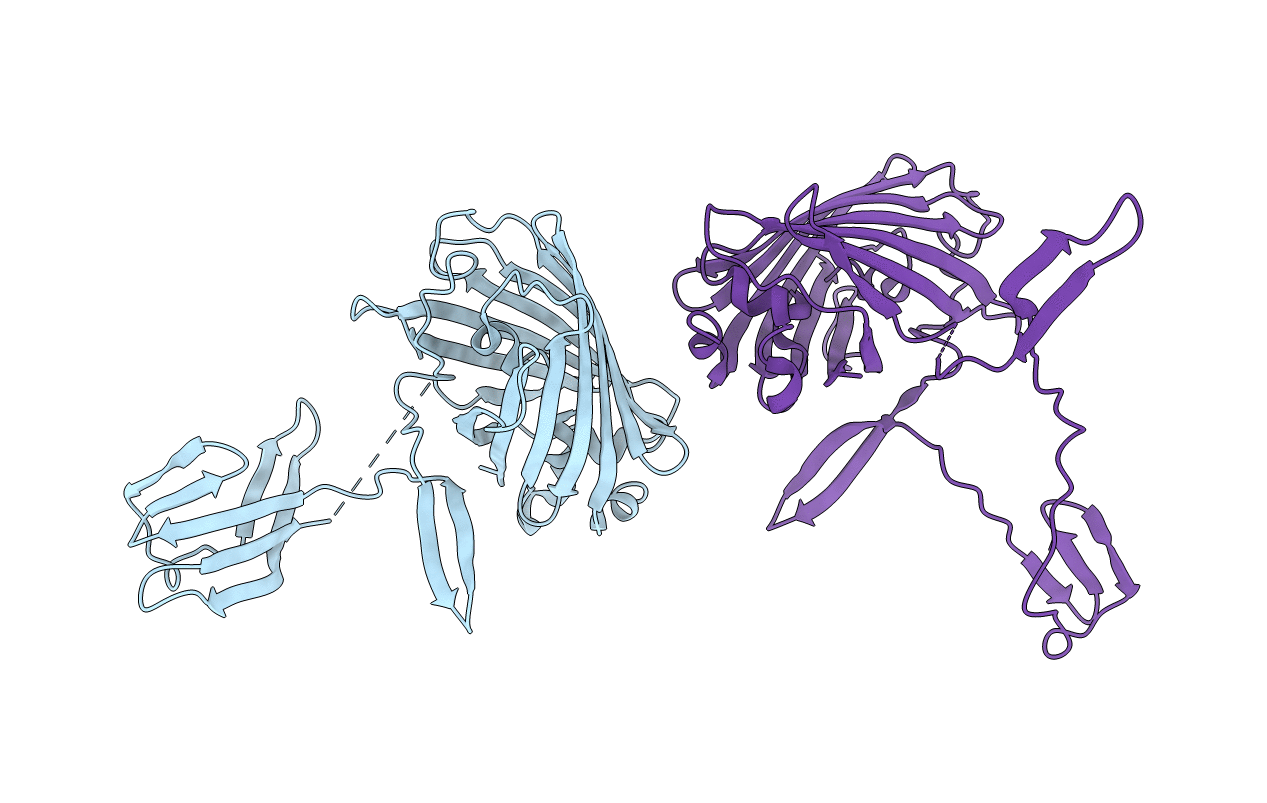
Crystal structure of the carboxy-terminal domain of a coronavirus M protein fused with a split GFP
Biological Source:
Source Organism(s):
Aequorea victoria (Taxon ID: 6100)
Pipistrellus bat coronavirus HKU5 (Taxon ID: 694008)
Pipistrellus bat coronavirus HKU5 (Taxon ID: 694008)
Expression System(s):
Method Details:
Experimental Method:
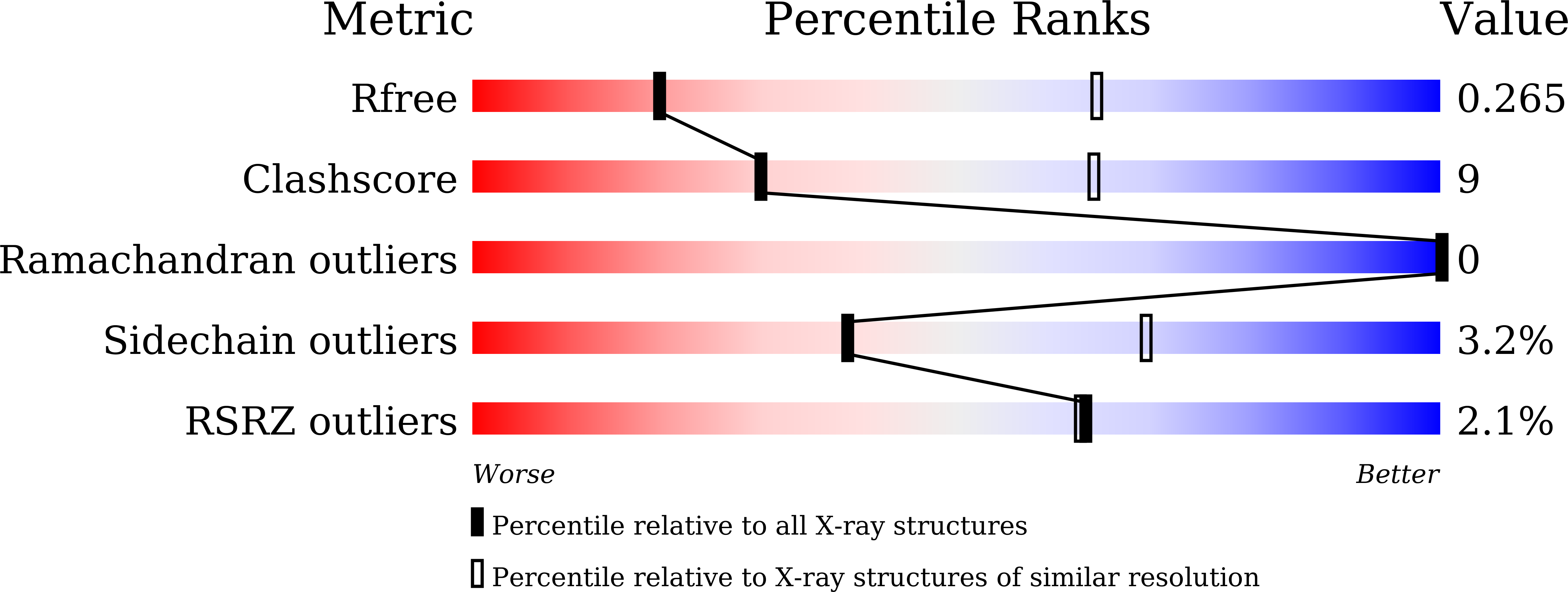
Resolution:
3.42 Å
R-Value Free:
0.26
R-Value Work:
0.23
R-Value Observed:
0.24
Space Group:
P 61 2 2