Abstact
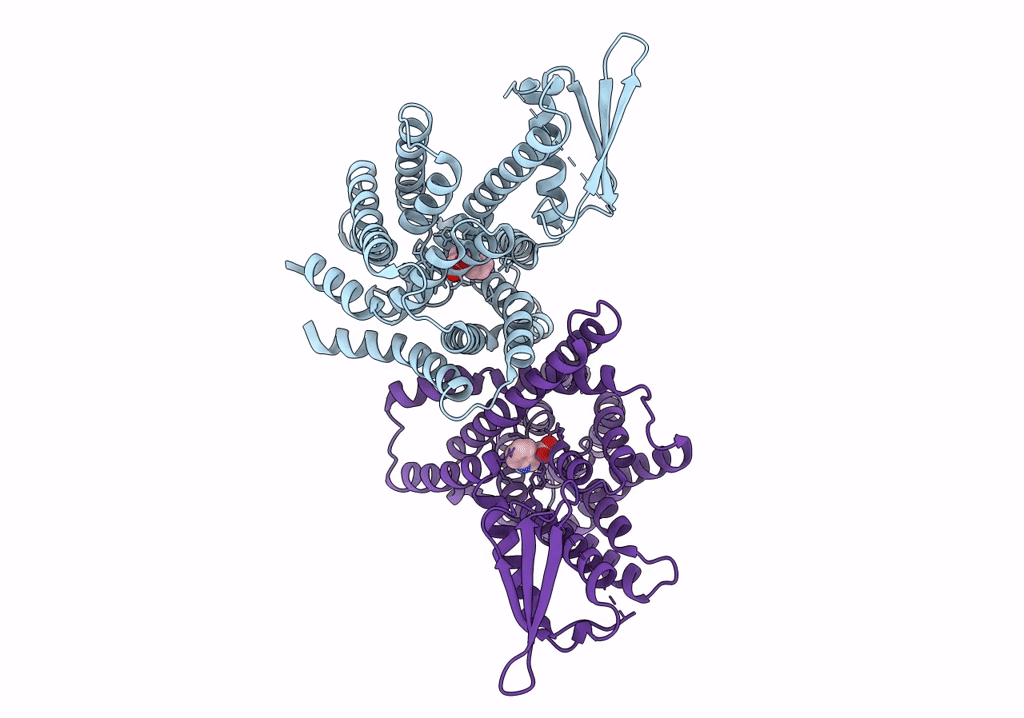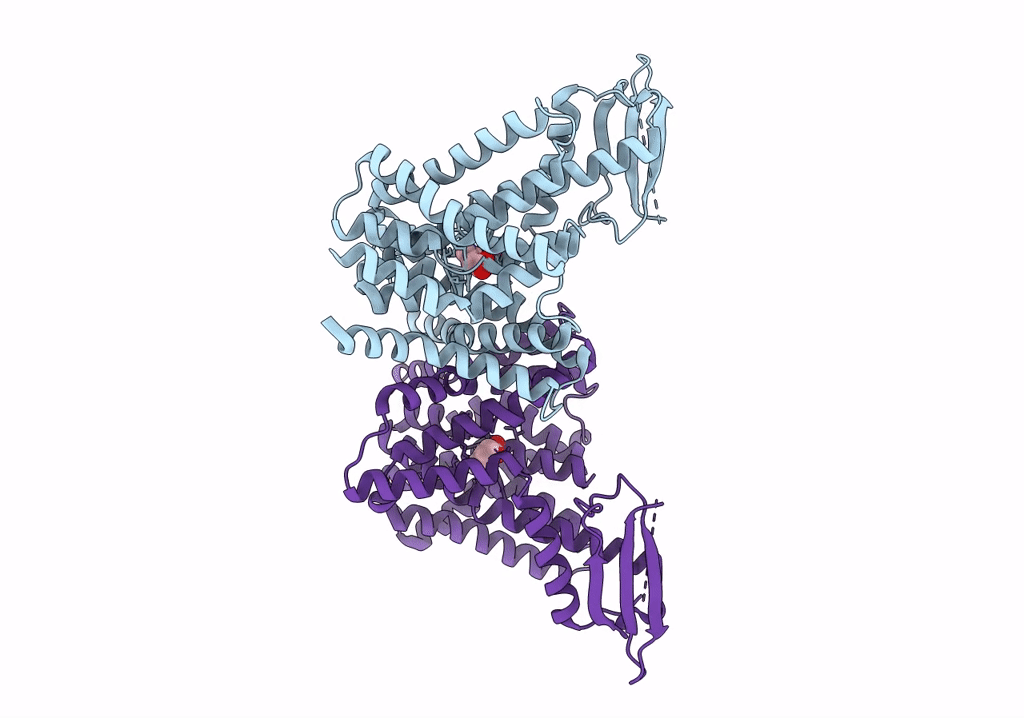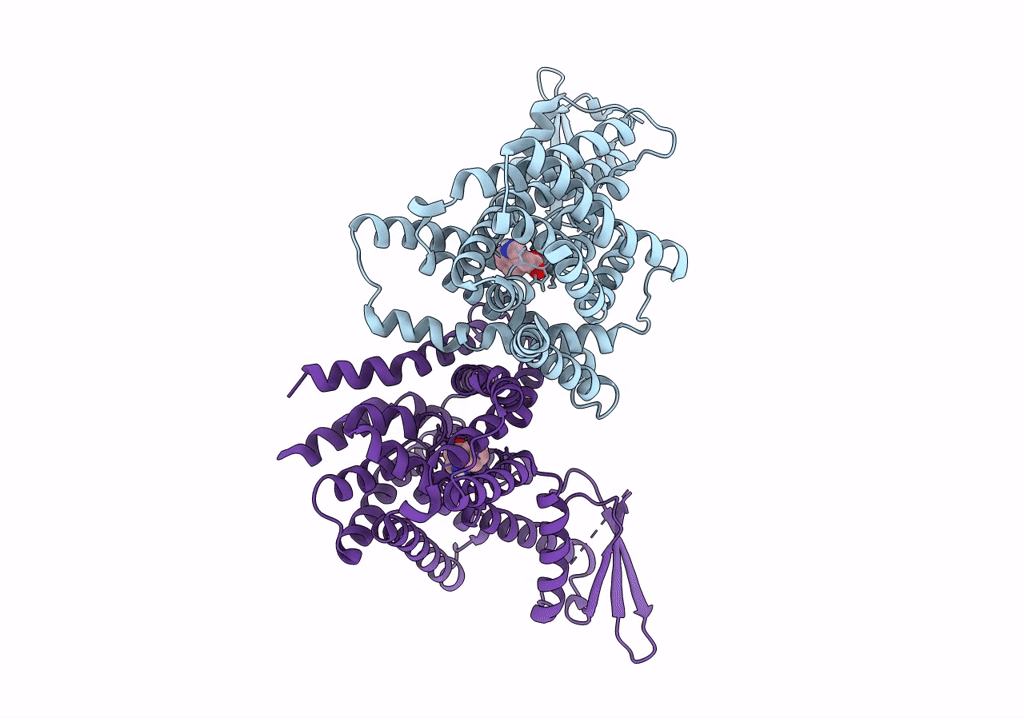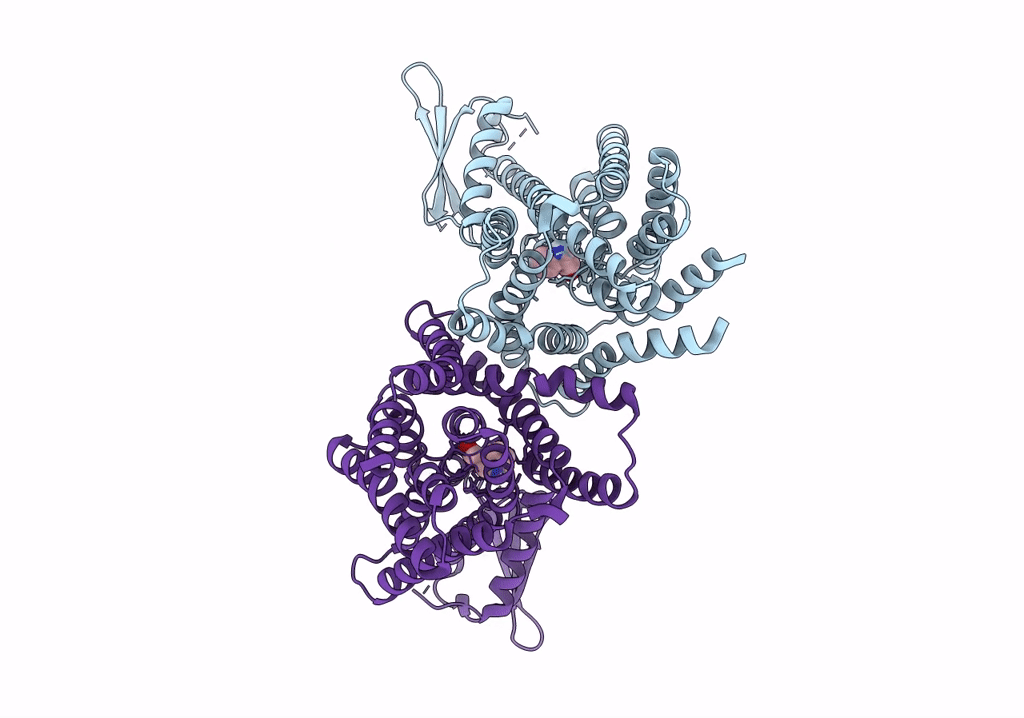
The PIN-FORMED (PIN) protein family of auxin transporters mediates polar auxin transport and has crucial roles in plant growth and development1,2. Here we present cryo-electron microscopy structures of PIN3 from Arabidopsis thaliana in the apo state and in complex with its substrate indole-3-acetic acid and the inhibitor N-1-naphthylphthalamic acid (NPA). A. thaliana PIN3 exists as a homodimer, and its transmembrane helices 1, 2 and 7 in the scaffold domain are involved in dimerization. The dimeric PIN3 forms a large, joint extracellular-facing cavity at the dimer interface while each subunit adopts an inward-facing conformation. The structural and functional analyses, along with computational studies, reveal the structural basis for the recognition of indole-3-acetic acid and NPA and elucidate the molecular mechanism of NPA inhibition on PIN-mediated auxin transport. The PIN3 structures support an elevator-like model for the transport of auxin, whereby the transport domains undergo up-down rigid-body motions and the dimerized scaffold domains remain static.