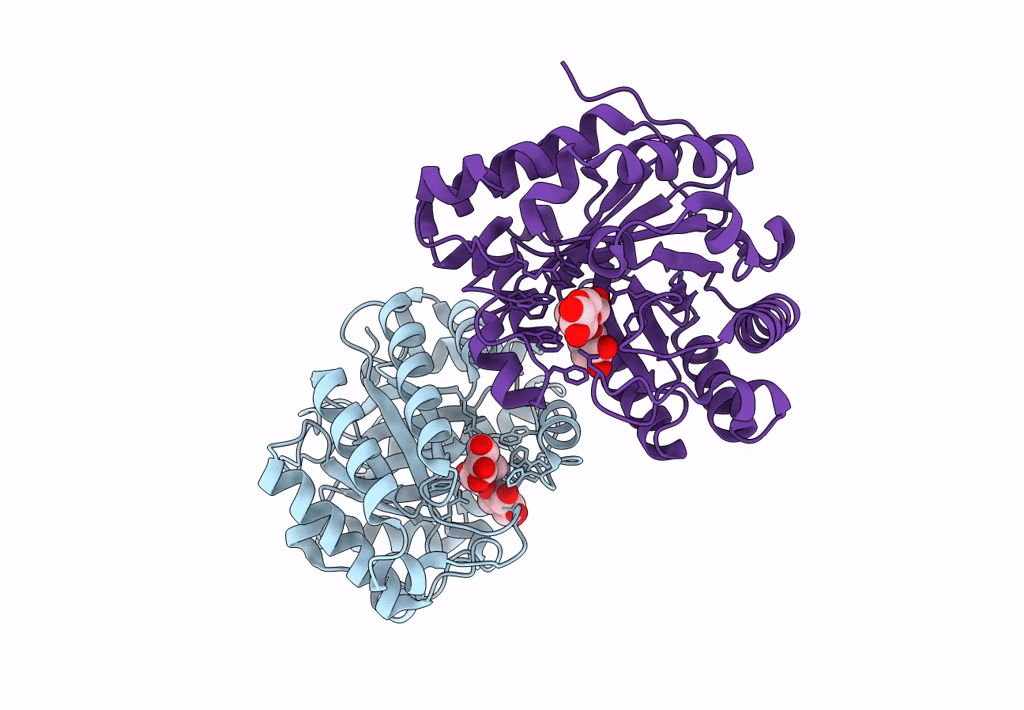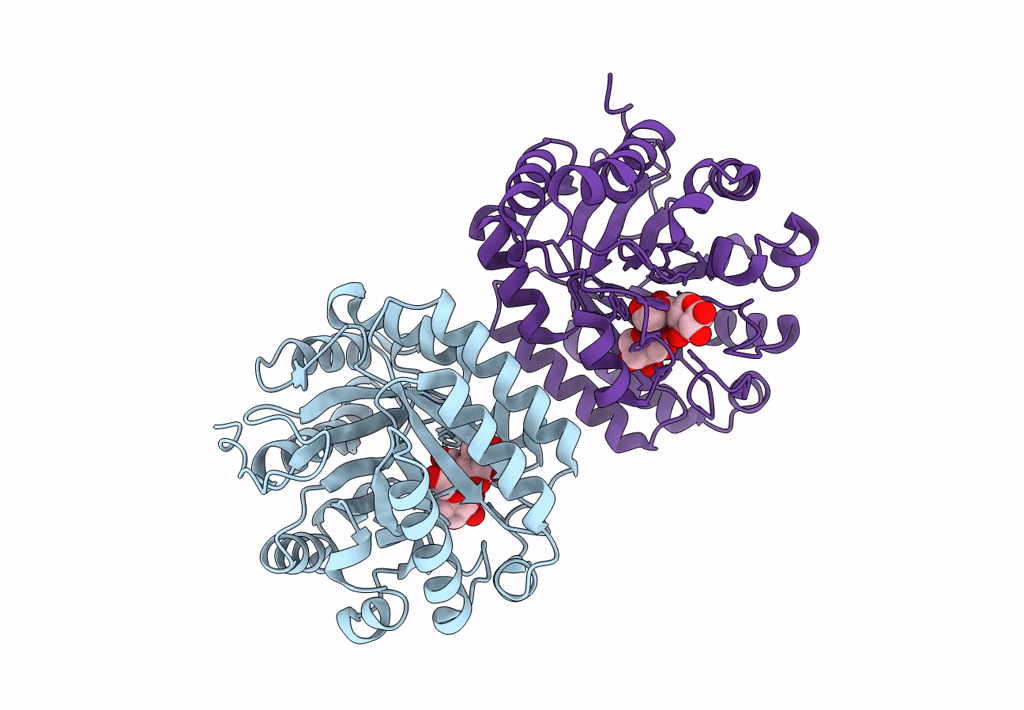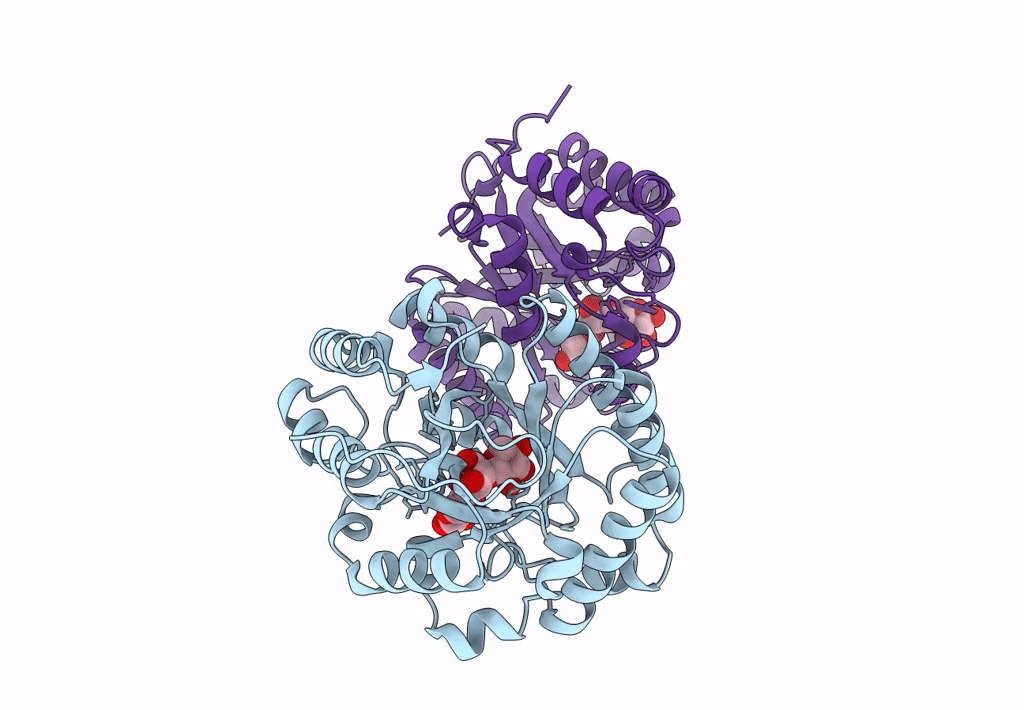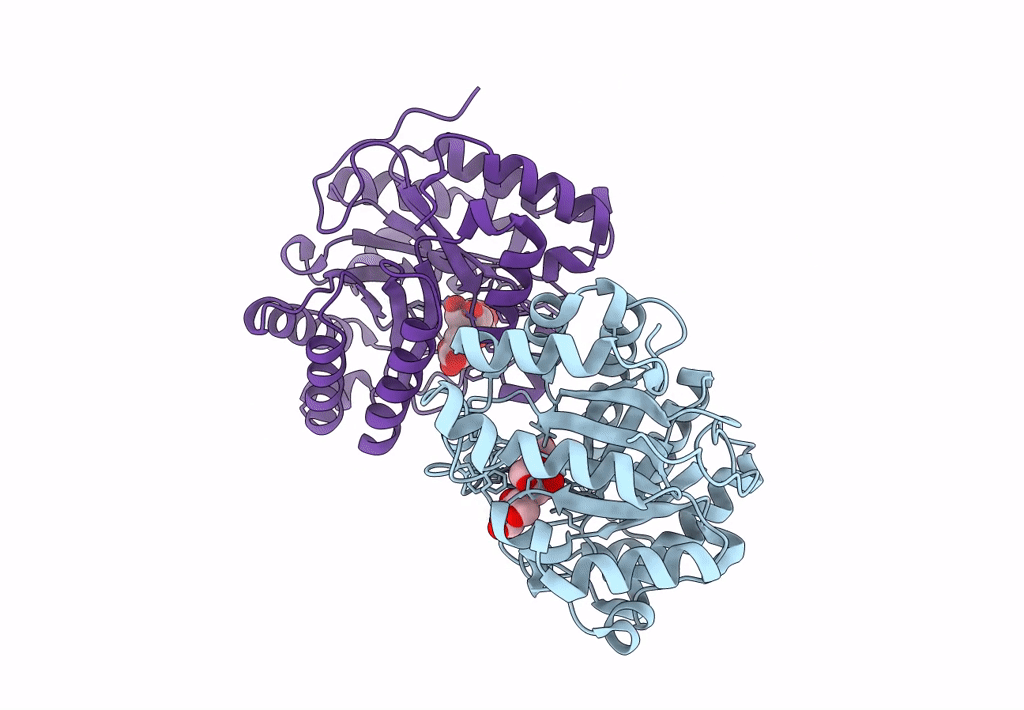
Abstact
Xylans are polysaccharides composed of xylose and include β1,4-xylan, β1,3-xylan, and β1,3/1,4-mixed-linkage xylan (MLX). MLX is widely present in marine red algae and constitutes a significant organic carbon in the ocean. Xylanases are hydrolase enzymes that play an important role in xylan degradation. While a variety of β1,4-xylanases and β1,3-xylanases involved in the degradation of β1,4-xylan and β1,3-xylan have been reported, no specific enzyme has yet been identified that degrades MLX. Herein, we report the characterization of a new MLX-specific xylanase from the marine bacterium Polaribacter sp. Q13 which utilizes MLX for growth. The bacterium secretes xylanases to degrade MLX, among which is Xyn26A, an MLX-specific xylanase that shows low sequence similarities (<27%) to β1,3-xylanases in the glycoside hydrolase family 26 (GH26). We show that Xyn26A attacks MLX precisely at β1,4-linkages, following a β1,3-linkage toward the reducing end. We confirm that Xyn26A and its homologs have the same specificity and mode of action on MLX, and thus represent a new xylanase group which we term as MLXases. We further solved the structure of a representative MLXase, AlXyn26A. Structural and biochemical analyses revealed that the specificity of MLXases depends critically on a precisely positioned β1,3-linkage at the -2/-1 subsite. Compared to the GH26 β1,3-xylanases, we found MLXases have evolved a tunnel-shaped cavity that is fine-tuned to specifically recognize and hydrolyze MLX. Overall, this study offers a foremost insight into MLXases, shedding light on the biochemical mechanism of bacterial degradation of MLX.