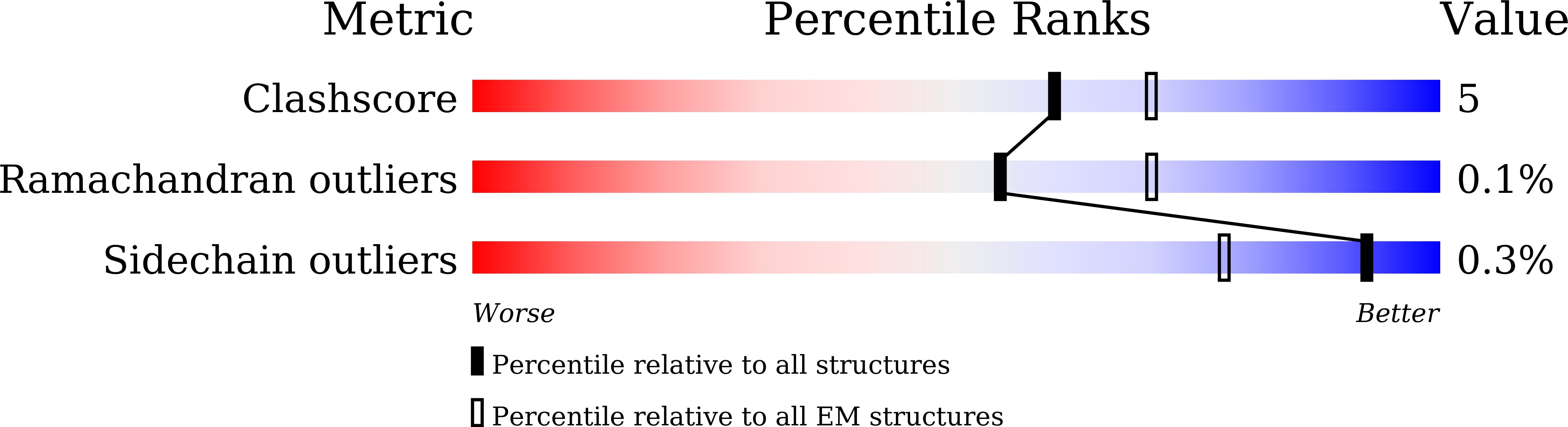
Deposition Date
2021-12-26
Release Date
2022-03-16
Last Version Date
2025-06-25
Entry Detail
PDB ID:
7WFF
Keywords:
Title:
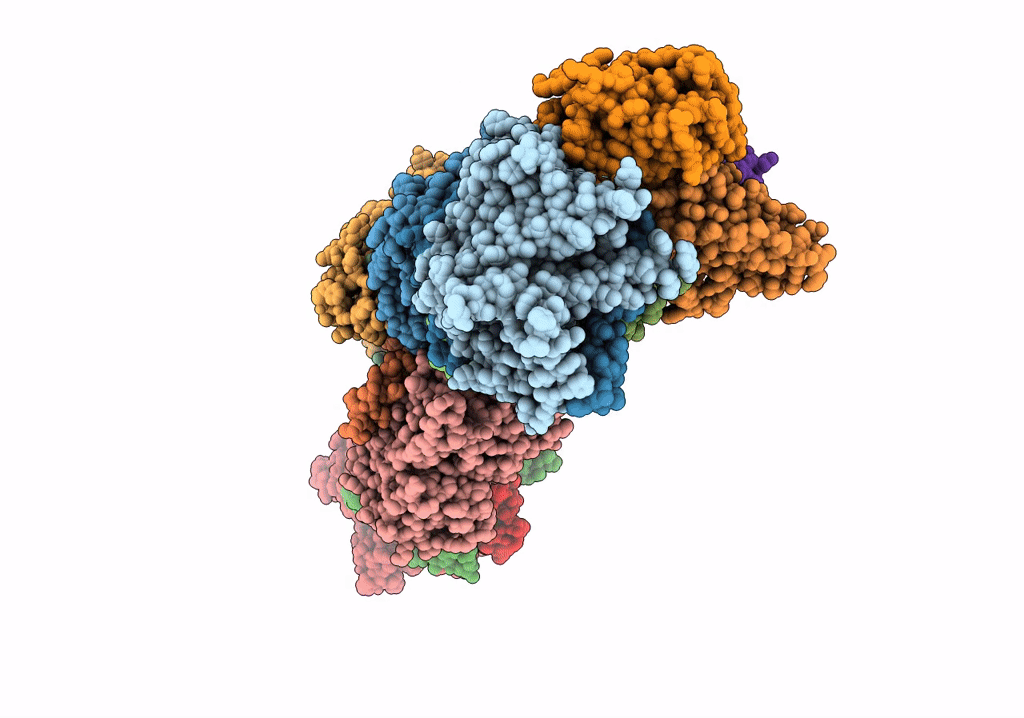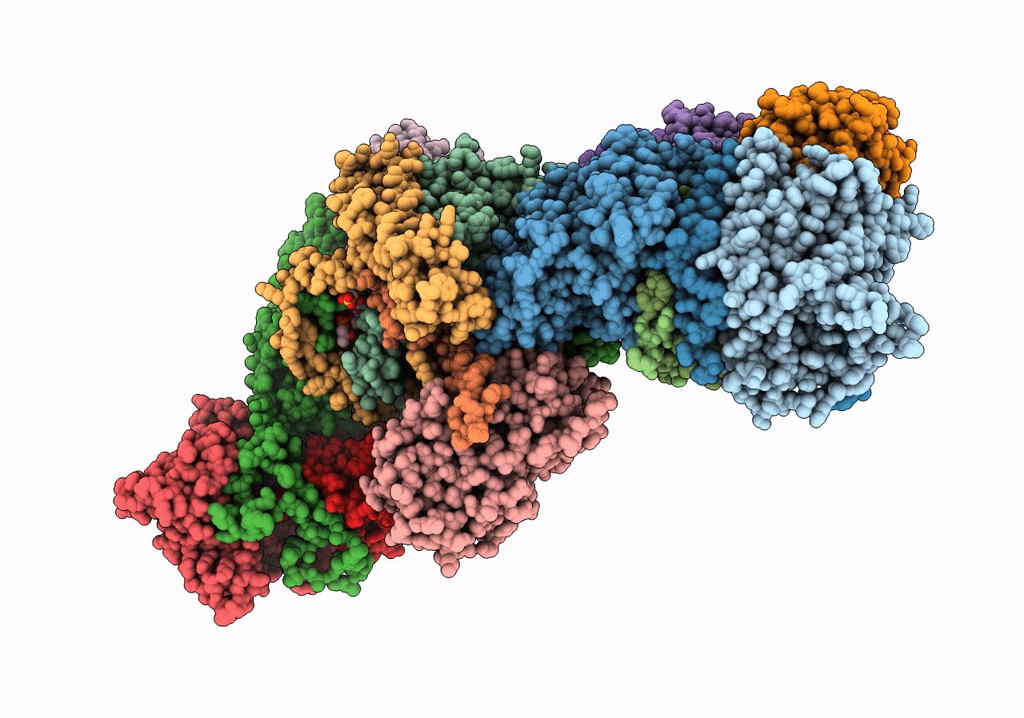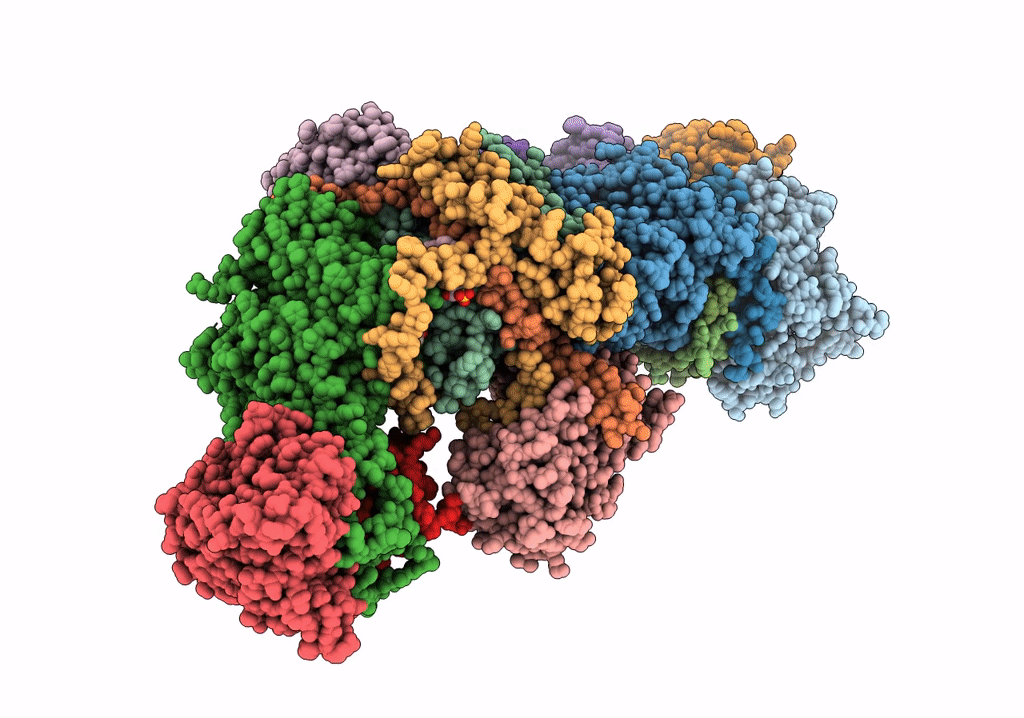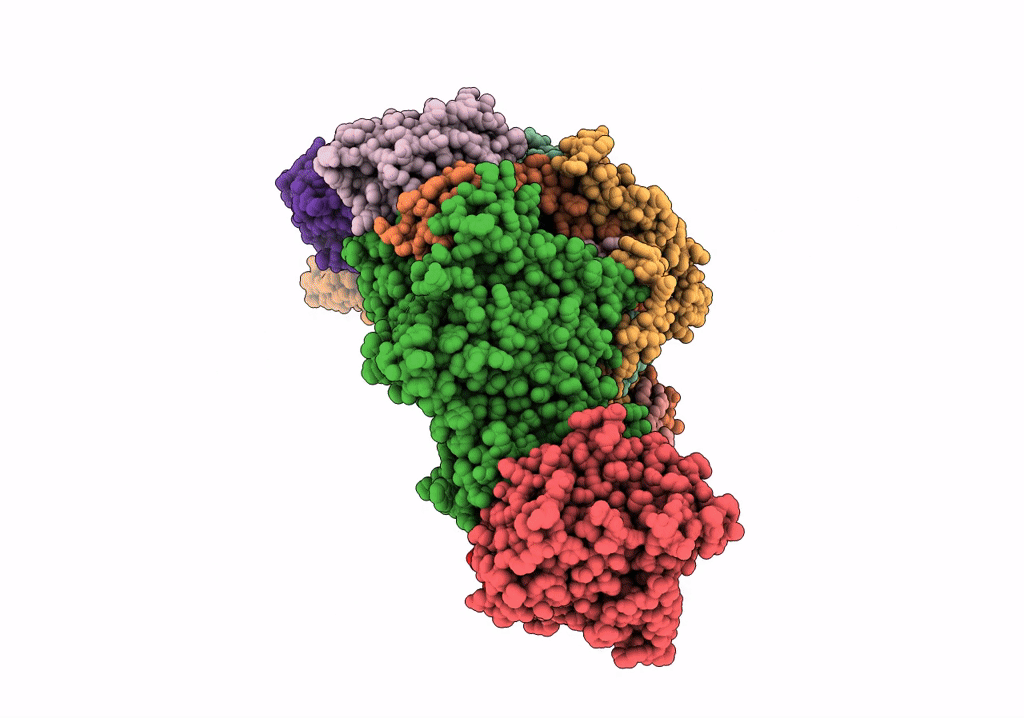
Subcomplexes B,M and L in the Cylic electron transfer supercomplex NDH-PSI from Arabidopsis
Biological Source:
Source Organism(s):
Arabidopsis thaliana (Taxon ID: 3702)
Method Details:
Experimental Method:
Resolution:
3.59 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE