Deposition Date
2021-12-17
Release Date
2023-07-05
Last Version Date
2025-06-25
Entry Detail
PDB ID:
7WBV
Keywords:
Title:
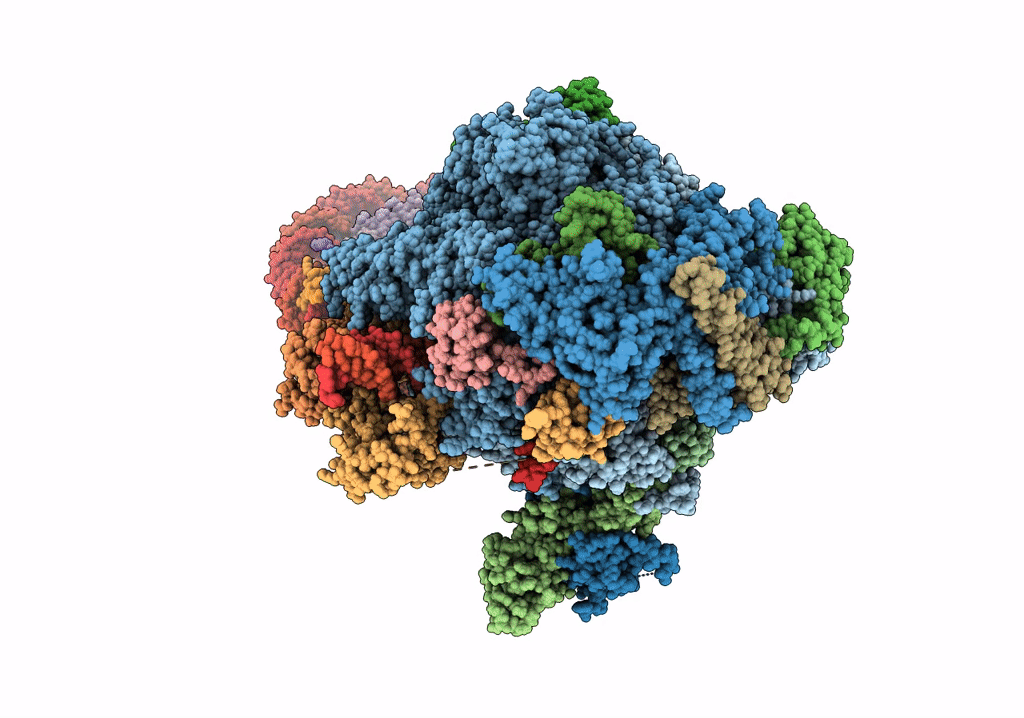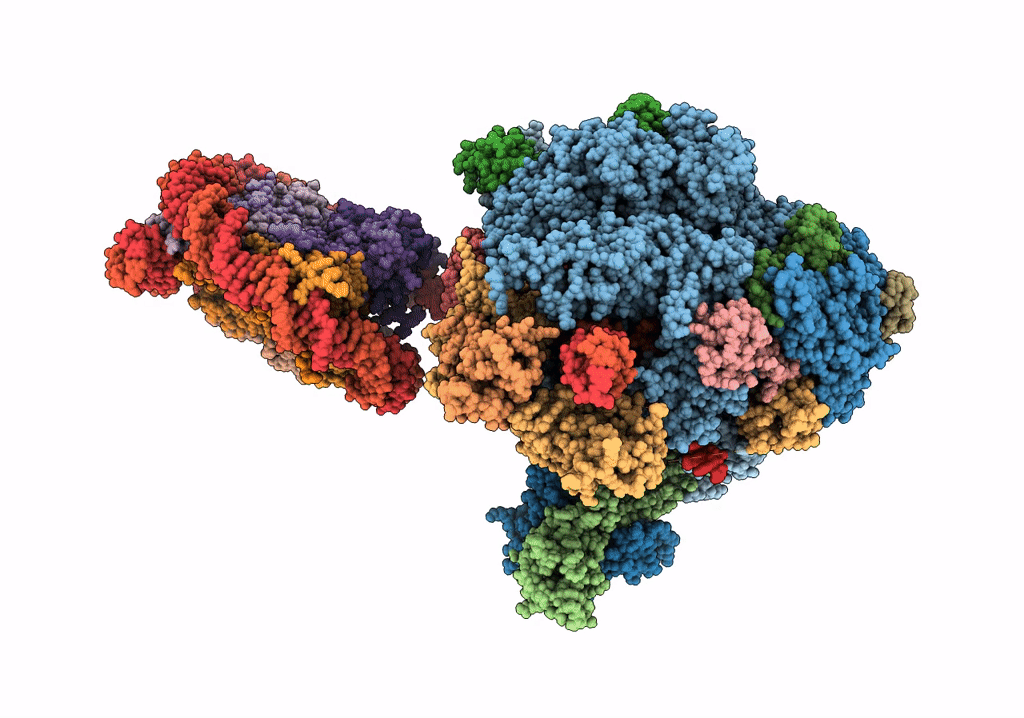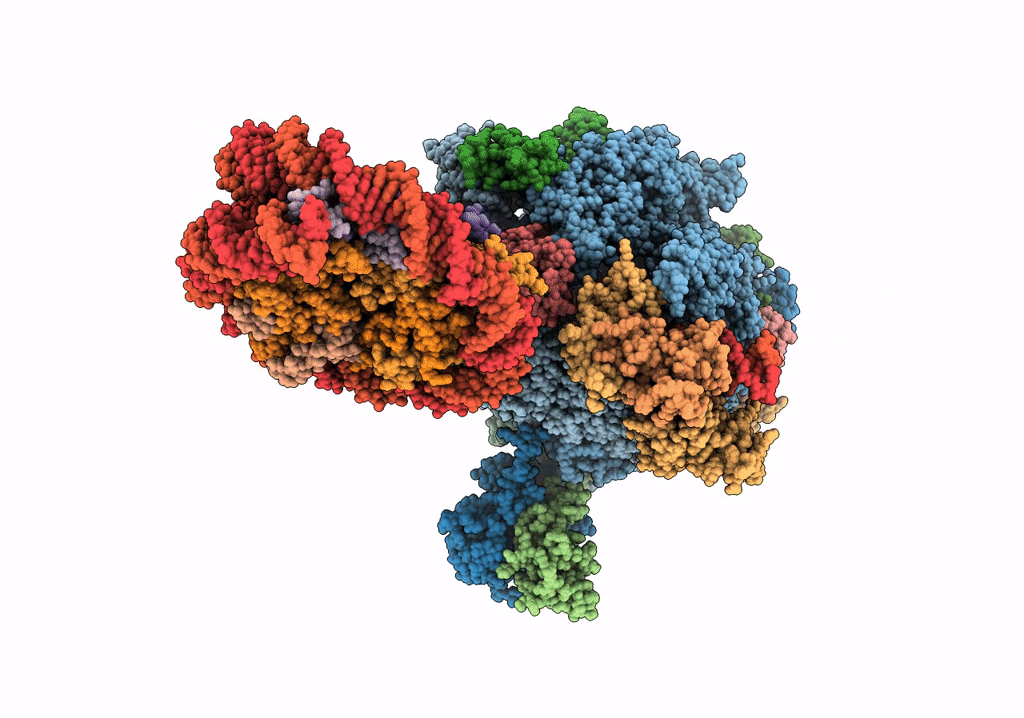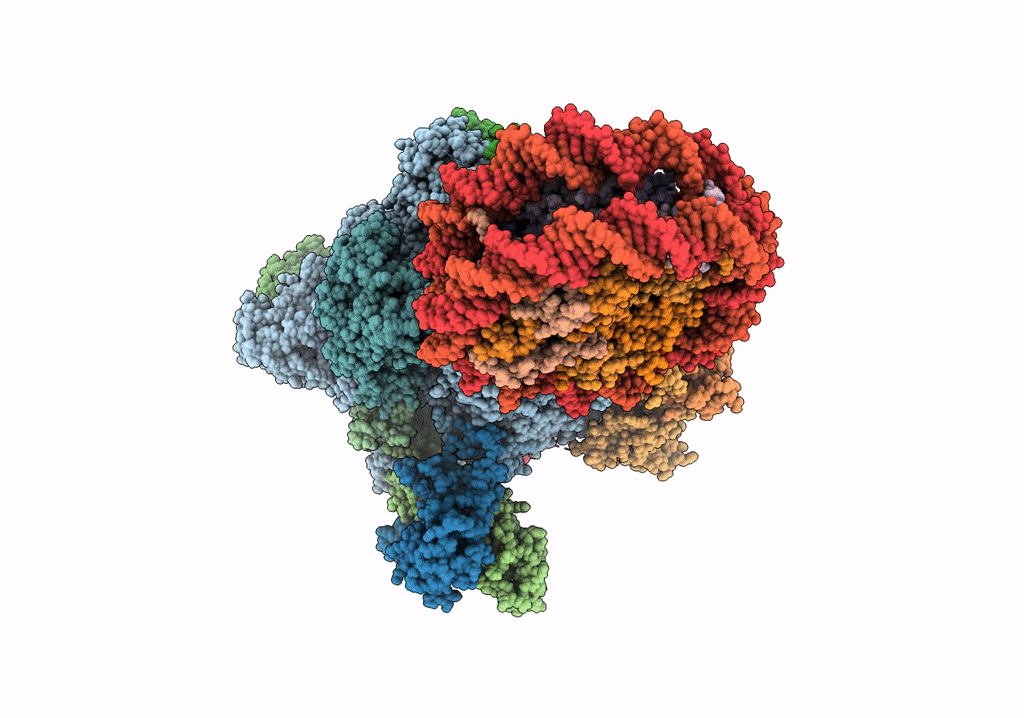
RNA polymerase II elongation complex bound with Elf1 and Spt4/5, stalled at SHL(-4) of the nucleosome
Biological Source:
Source Organism(s):
Komagataella phaffii (Taxon ID: 460519)
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Homo sapiens (Taxon ID: 9606)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.10 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


