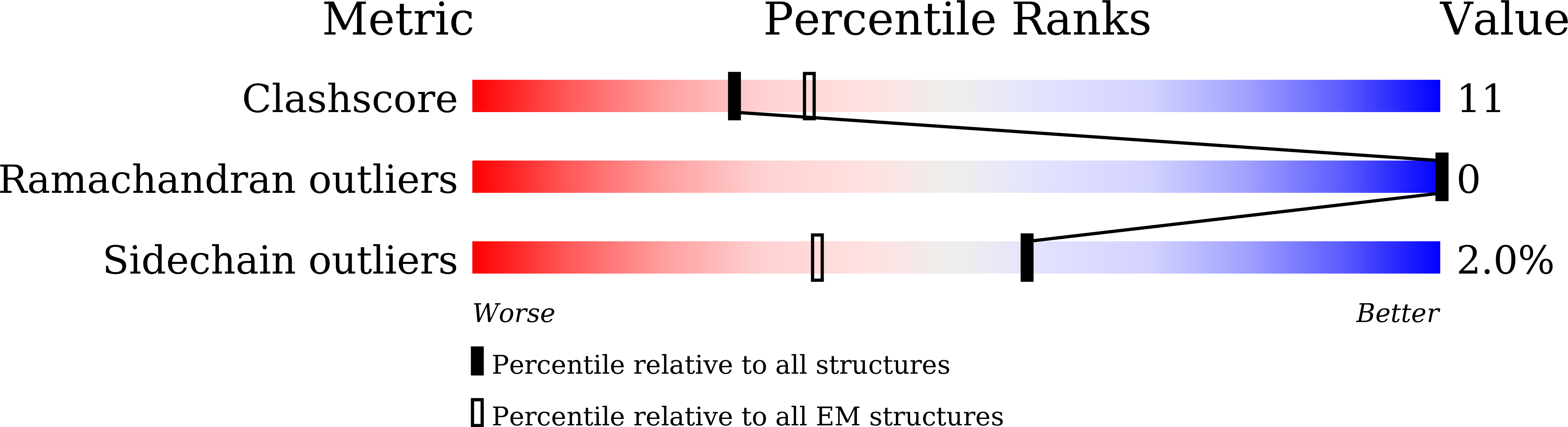
Deposition Date
2021-11-13
Release Date
2022-04-27
Last Version Date
2024-11-13
Entry Detail
PDB ID:
7VY3
Keywords:
Title:
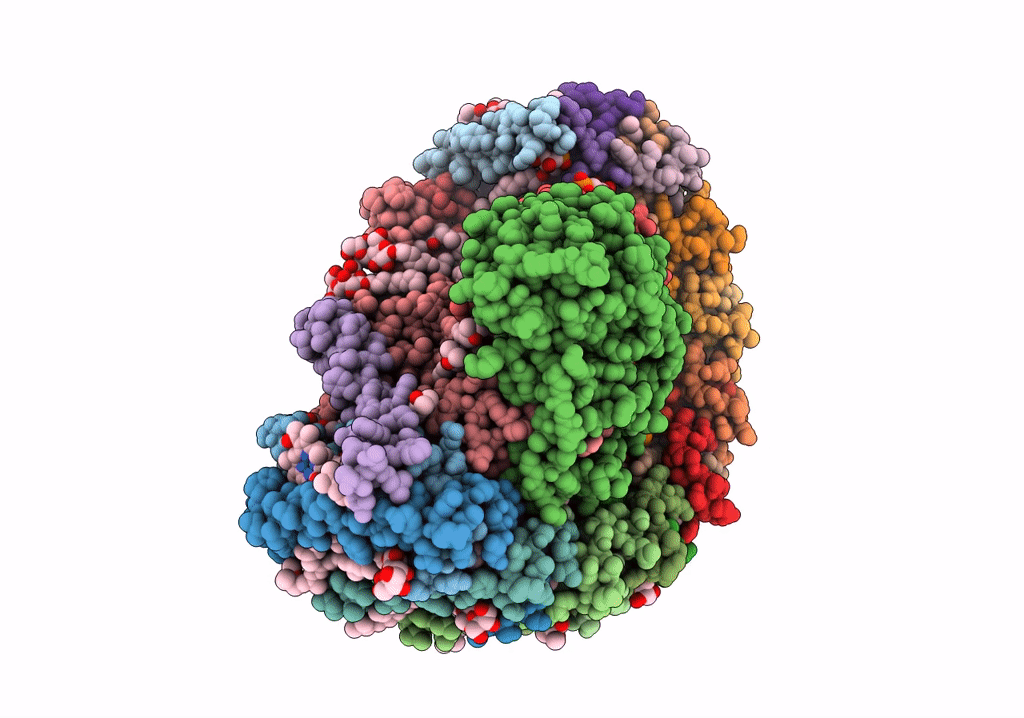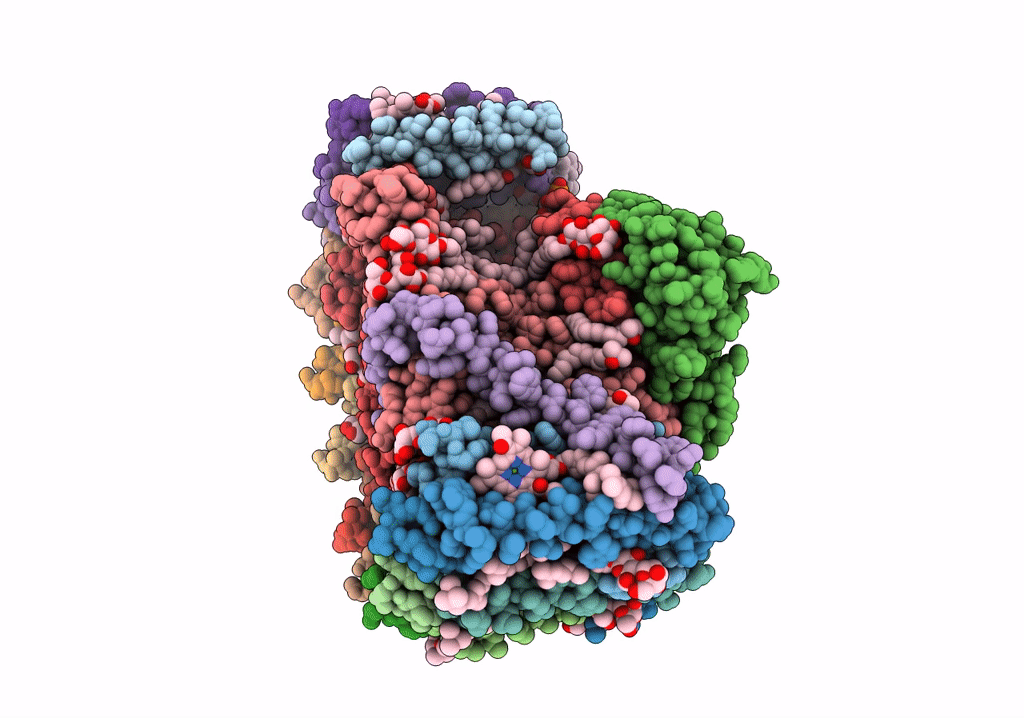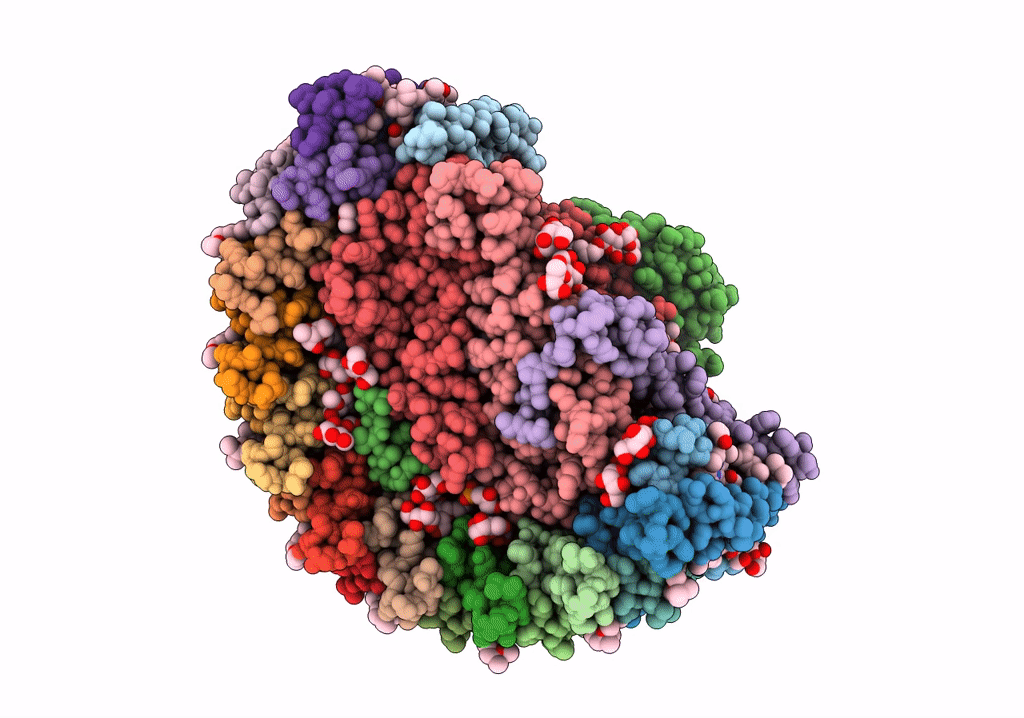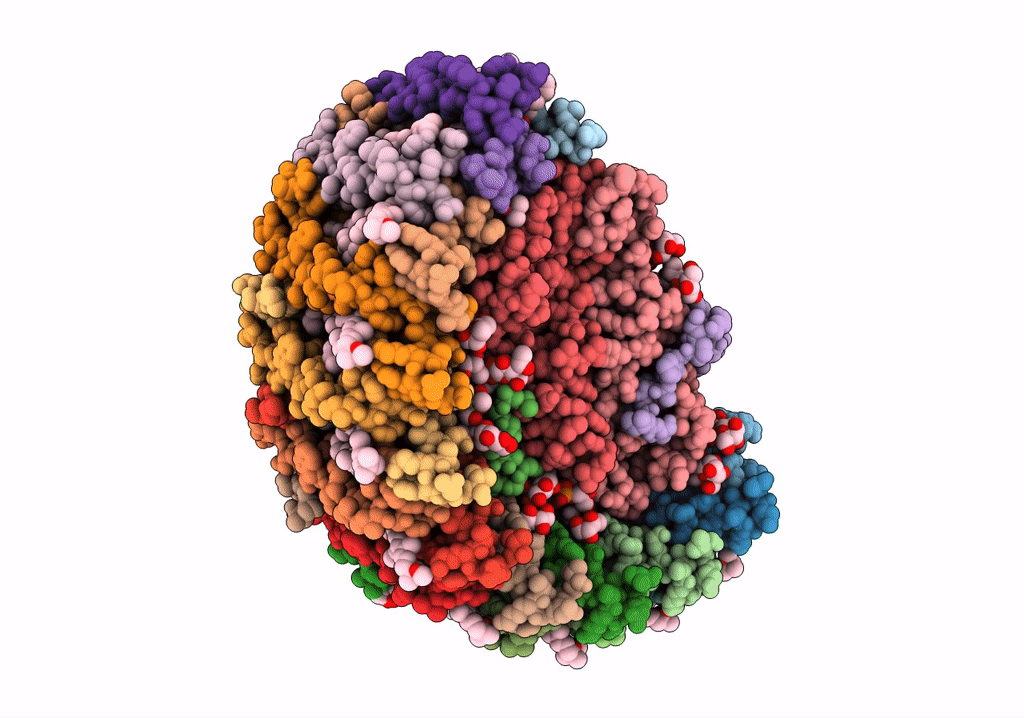
STRUCTURE OF PHOTOSYNTHETIC LH1-RC SUPER-COMPLEX OF RHODOBACTER SPHAEROIDES LACKING PROTEIN-U
Biological Source:
Source Organism(s):
Rhodobacter sphaeroides f. sp. denitrificans (Taxon ID: 39723)
Method Details:
Experimental Method:
Resolution:
2.63 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE