Deposition Date
2021-11-01
Release Date
2022-06-01
Last Version Date
2024-04-03
Entry Detail
PDB ID:
7VUA
Keywords:
Title:
Anaerobic hydroxyproline degradation involving C-N cleavage by a glycyl radical enzyme
Biological Source:
Source Organism:
Clostridiales bacterium (Taxon ID: 1898207)
Host Organism:
Method Details:
Experimental Method:
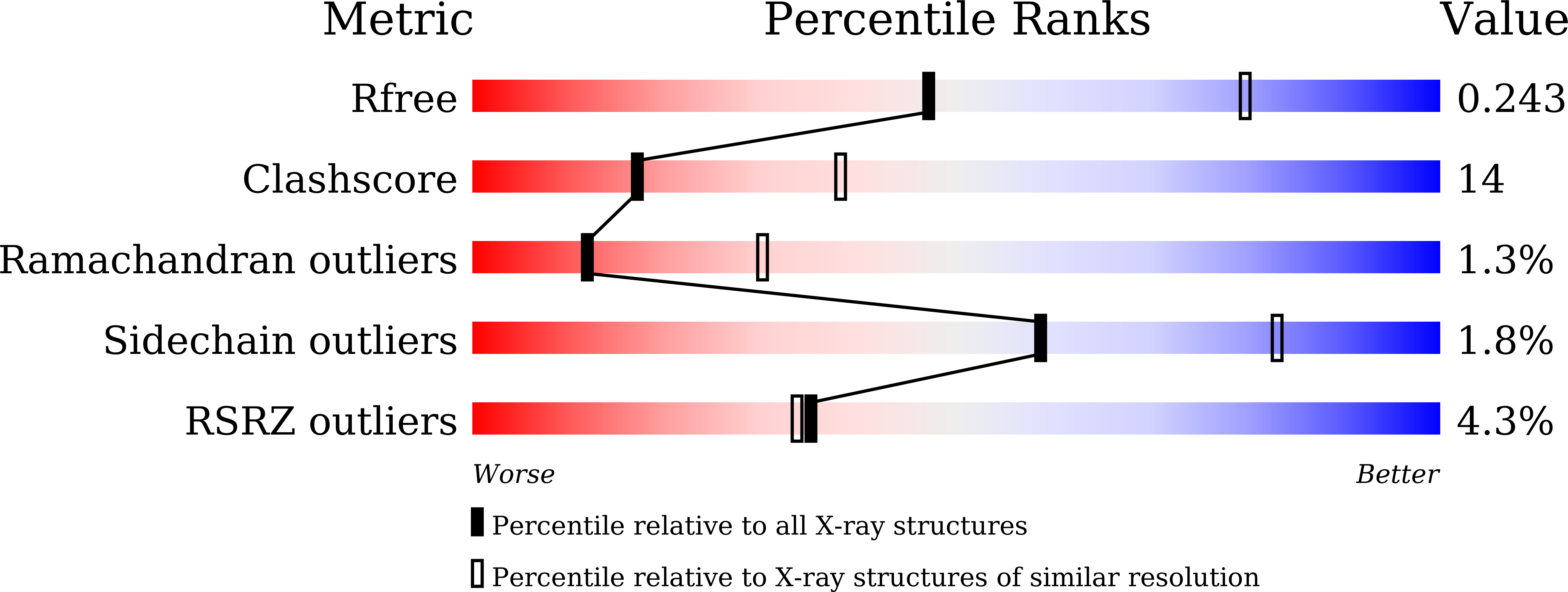
Resolution:
2.70 Å
R-Value Free:
0.24
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
C 2 2 21