Deposition Date
2021-08-06
Release Date
2021-11-03
Last Version Date
2023-11-29
Entry Detail
PDB ID:
7V1V
Keywords:
Title:
Difructose dianhydride I synthase/hydrolase (alphaFFase1) from Bifidobacterium dentium, ligand-free form
Biological Source:
Source Organism(s):
Bifidobacterium dentium (Taxon ID: 1689)
Expression System(s):
Method Details:
Experimental Method:
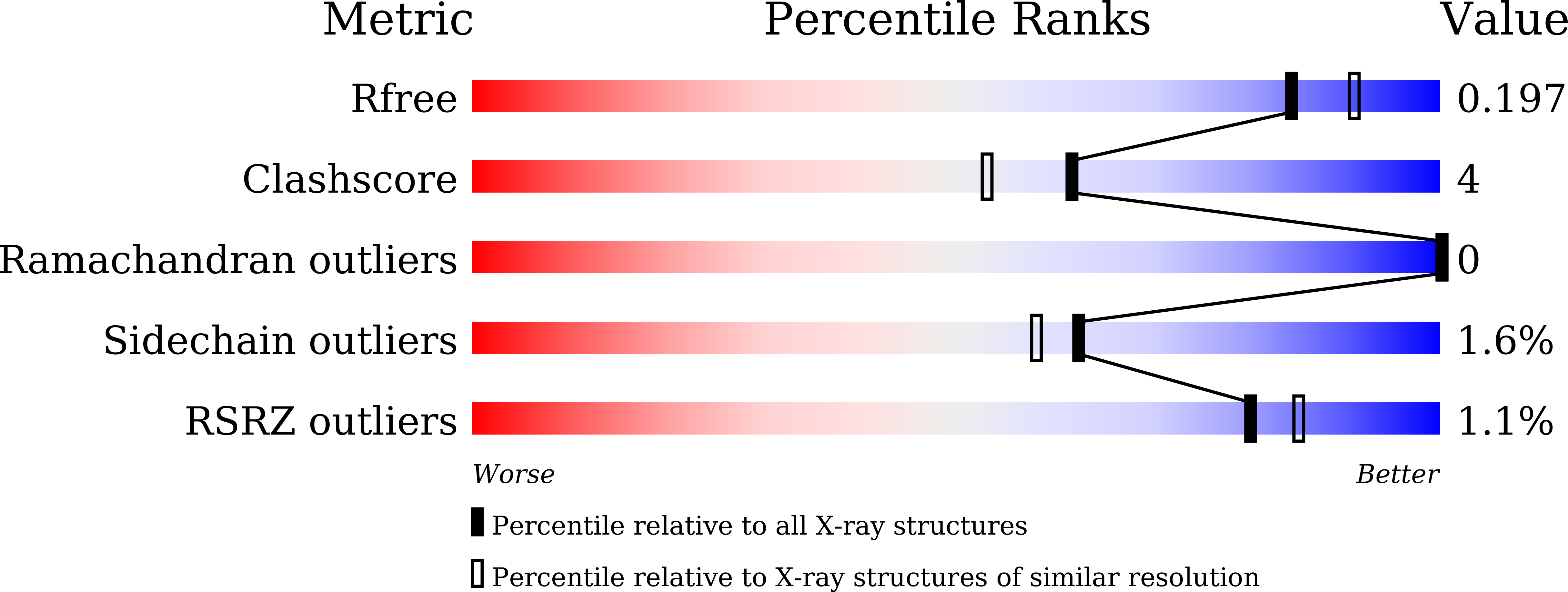
Resolution:
1.96 Å
R-Value Free:
0.19
R-Value Work:
0.15
R-Value Observed:
0.15
Space Group:
P 1 21 1