Deposition Date
2022-04-13
Release Date
2022-07-06
Last Version Date
2024-02-14
Entry Detail
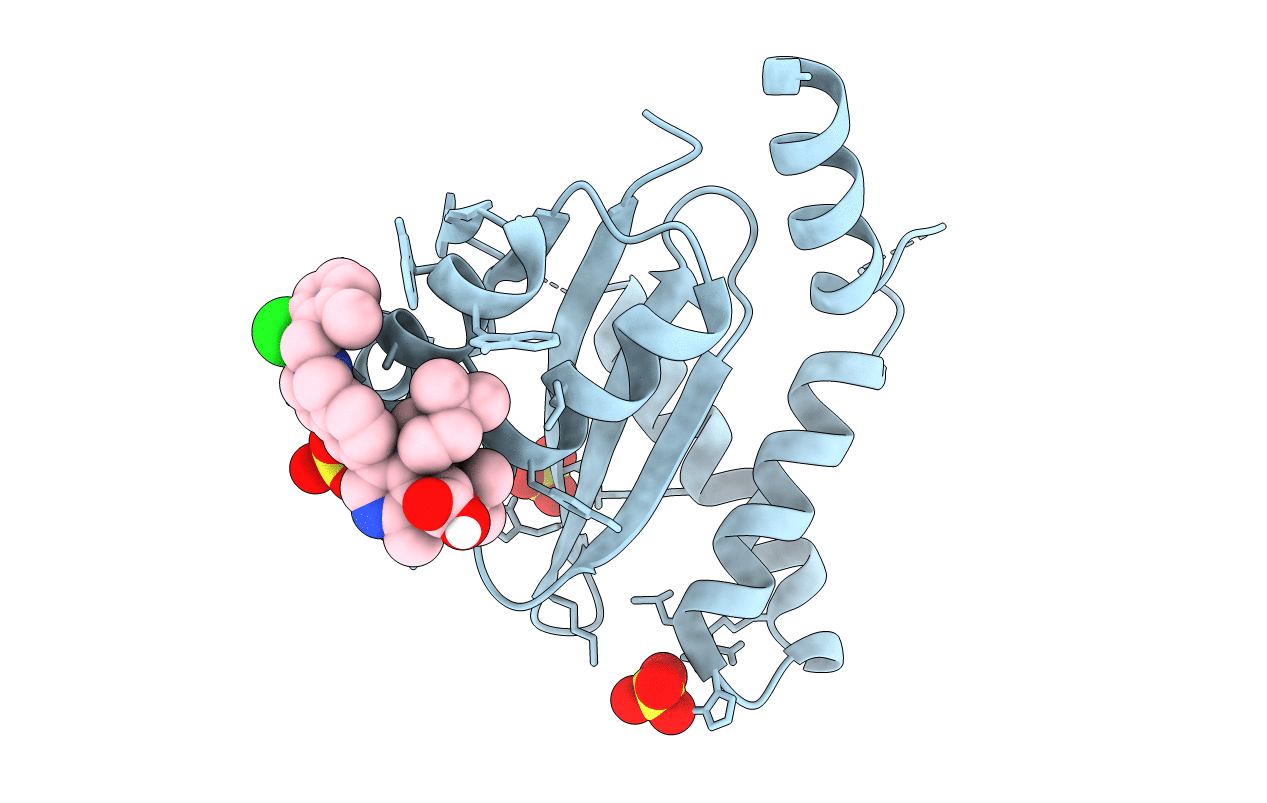
PDB ID:
7UOQ
Keywords:
Title:
CRYSTAL STRUCTURE OF HIV-1 INTEGRASE COMPLEXED WITH (2S)-2-(TERT-BUTOXY)-2-(5-{2-[(2-CHLORO-6-M ETHYLPHENYL)METHYL]-1,2,3,4-TETRAHYDROISOQUINOLIN-6-YL}-4- (4,4-DIMETHYLPIPERIDIN-1-YL)-2-METHYLPYRIDIN-3-YL)ACETIC ACID
Biological Source:
Source Organism:
Human immunodeficiency virus 1 (Taxon ID: 11676)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.89 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 31 2 1


