Abstact
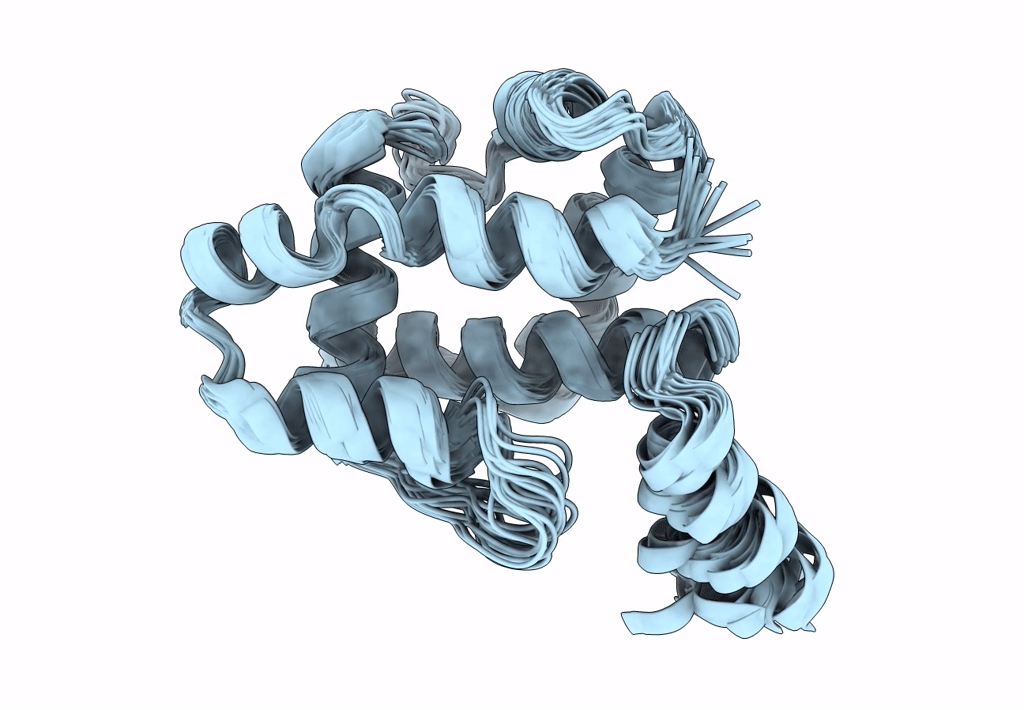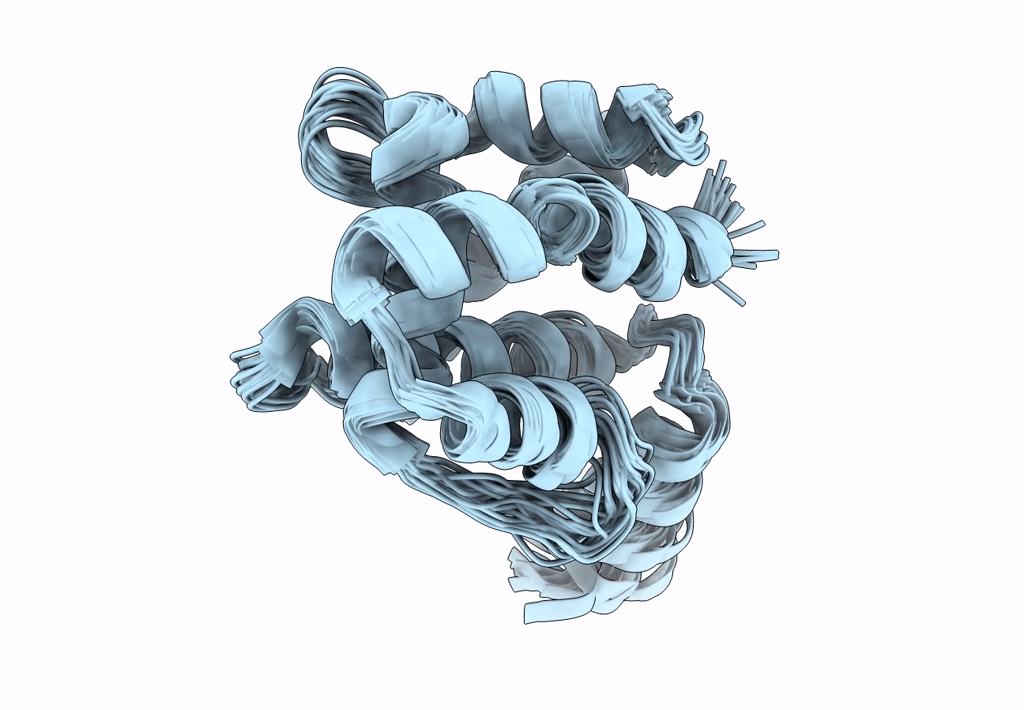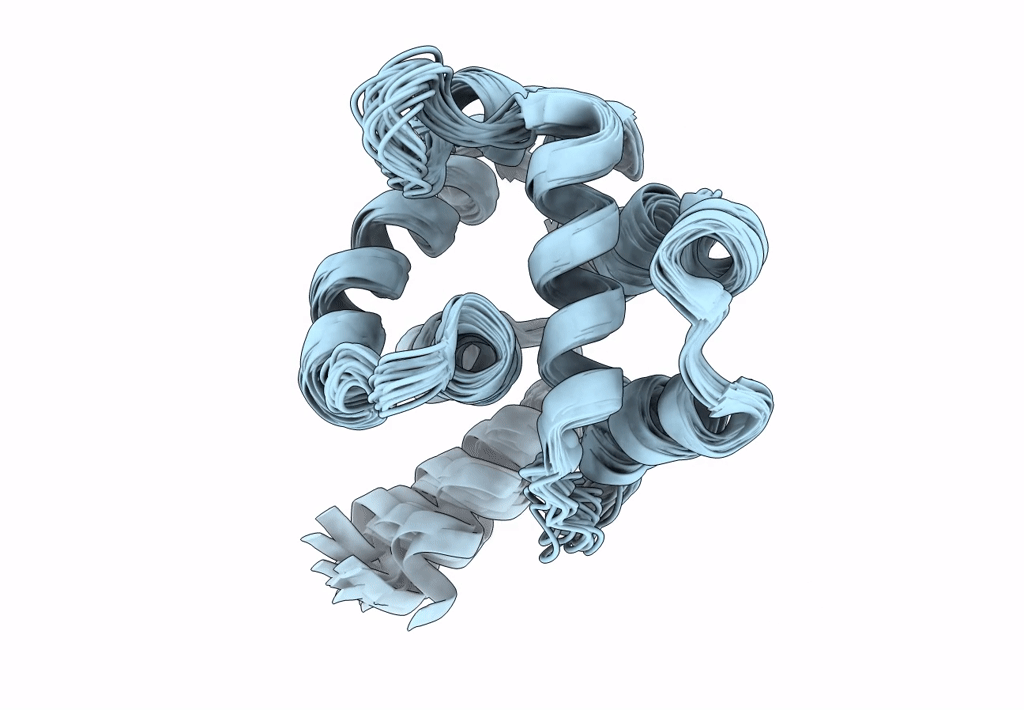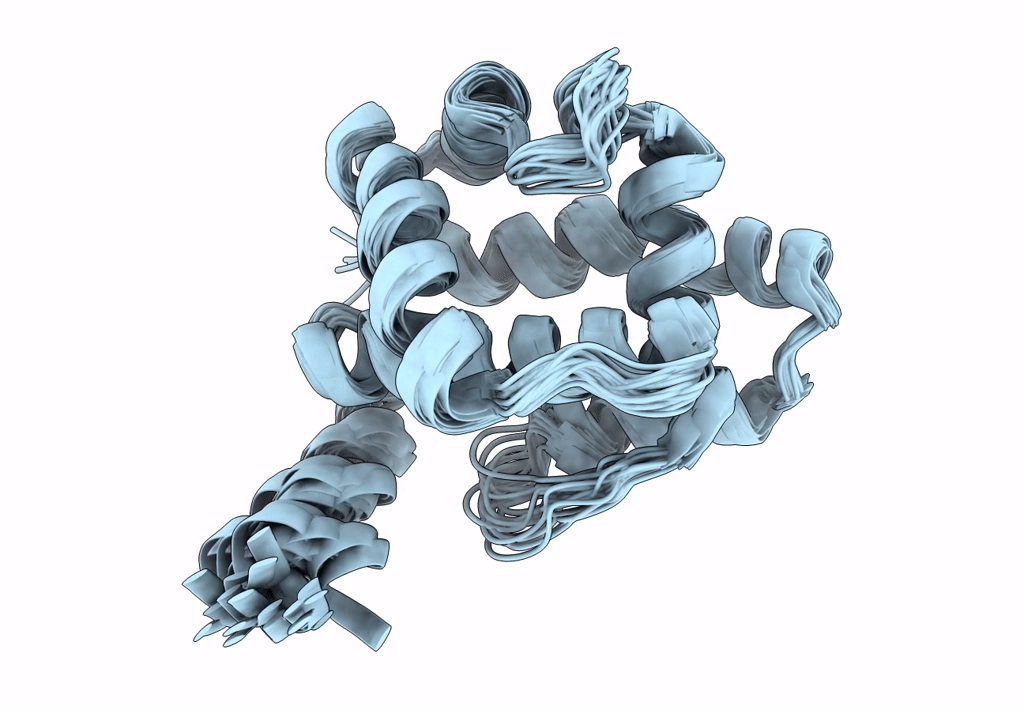
Ostrinia furnacalis is an invasive lepidopteran agricultural pest that relies on olfaction for mating and reproduction. Male moths have an extremely sensitive olfactory system that can detect the sex pheromones emitted by females over a great distance. Pheromone-binding proteins present in the male moth antenna play a key role in the pheromone uptake, transport, and release at the dendritic membrane of the olfactory neuron. Here, we report the first high-resolution NMR structure of a pheromone-binding protein from an Ostrinia species at pH 6.5. The core of the Ostrinia furnacalis PBP2 (OfurPBP2) consists of six helices, α1a (2-14), α1b (16-22), α2 (27-37), α3 (46-60), α4 (70-80), α5 (84-100), and α6 (107-124) surrounding a large hydrophobic pocket. The structure is stabilized by three disulfide bridges, 19-54, 50-108, and 97-117. In contrast to the unstructured C-terminus of other lepidopteran PBPs, the C-terminus of OfurPBP2 folds into an α-helix (α7) at pH 6.5. The protein has nanomolar affinity towards both pheromone isomers. Molecular docking of both pheromones, E-12 and Z-12-tetradecenyl acetate, to OfurPBP2 revealed that the residues Met5, Lys6, Met8, Thr9, Phe12, Phe36, Trp37, Phe76, Ser115, Phe118, Lys119, Ile122, His123, and Ala128 interact with both isomers, while Thr9 formed a hydrogen bond with the acetate head group. NMR structure and thermal unfolding studies with CD suggest that ligand release at pH 4.5 is likely due to the partial unfolding of the protein.