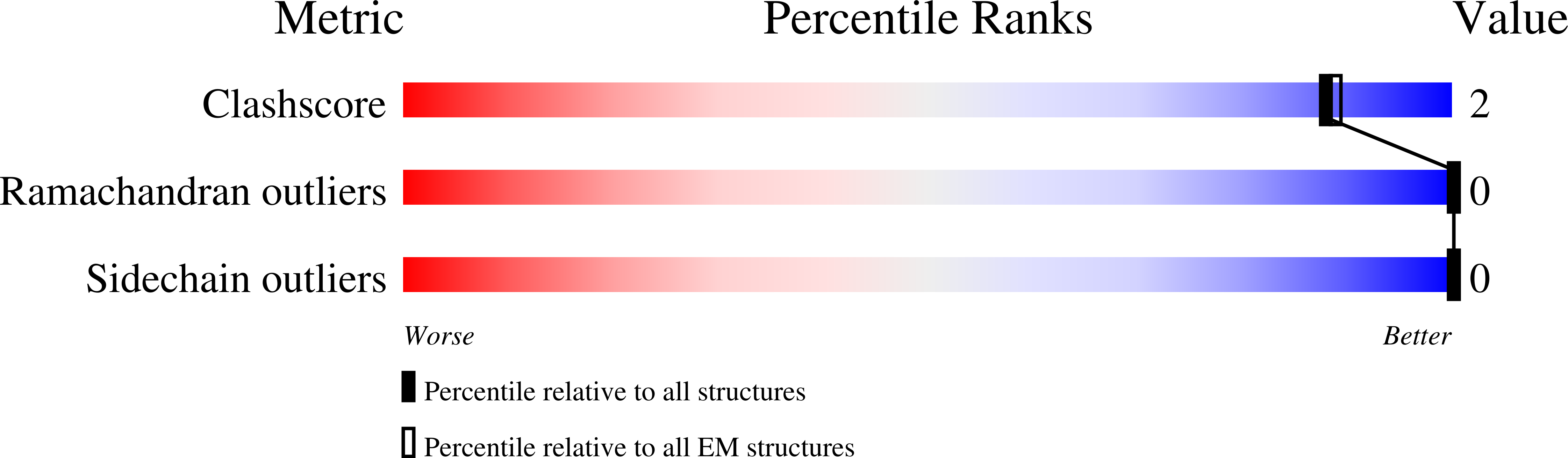
Deposition Date
2022-03-11
Release Date
2022-08-31
Last Version Date
2024-06-12
Entry Detail
PDB ID:
7UAA
Keywords:
Title:
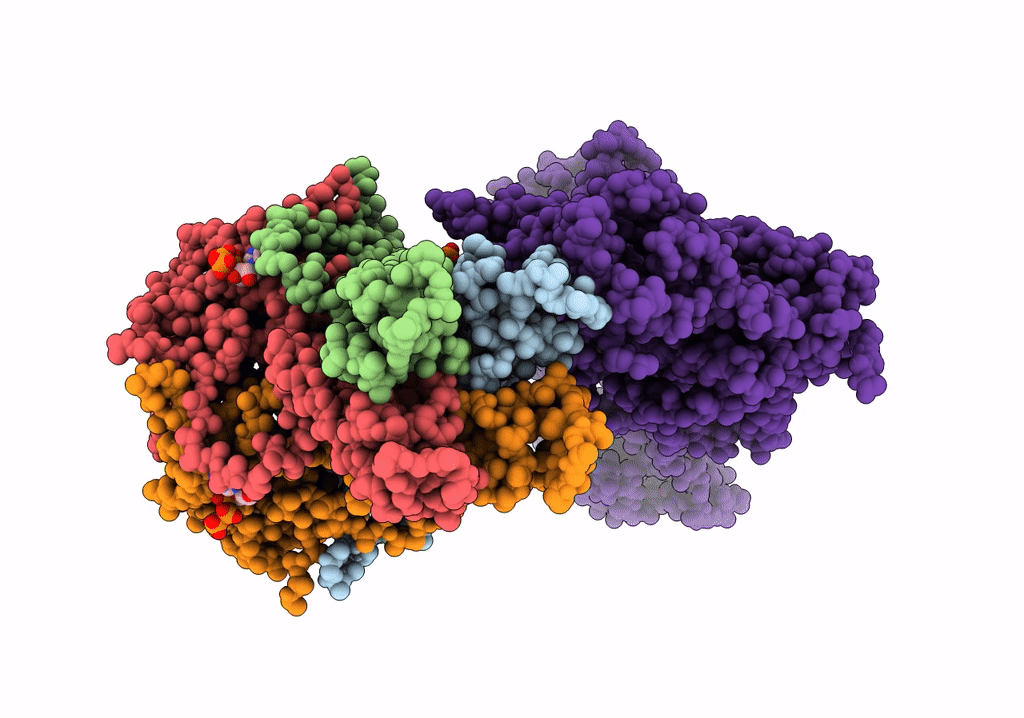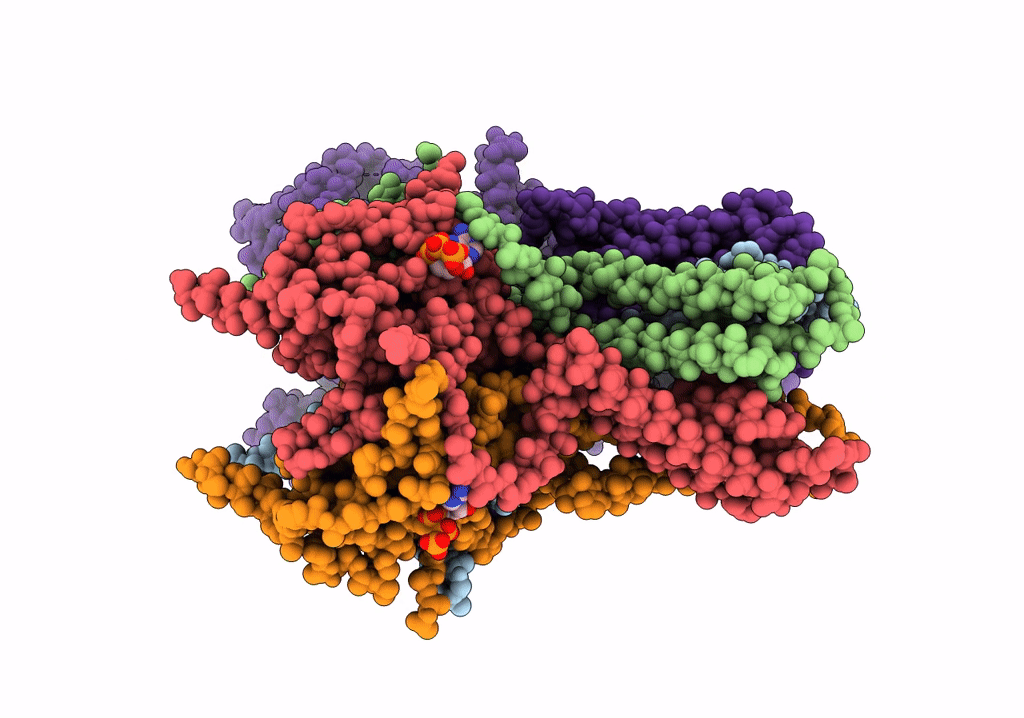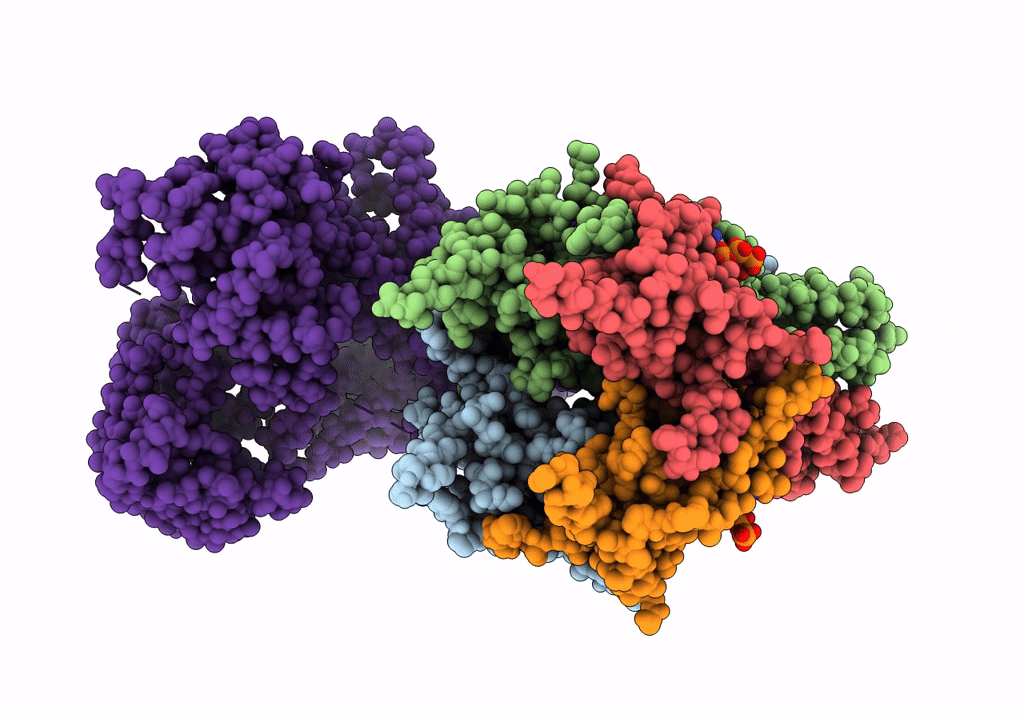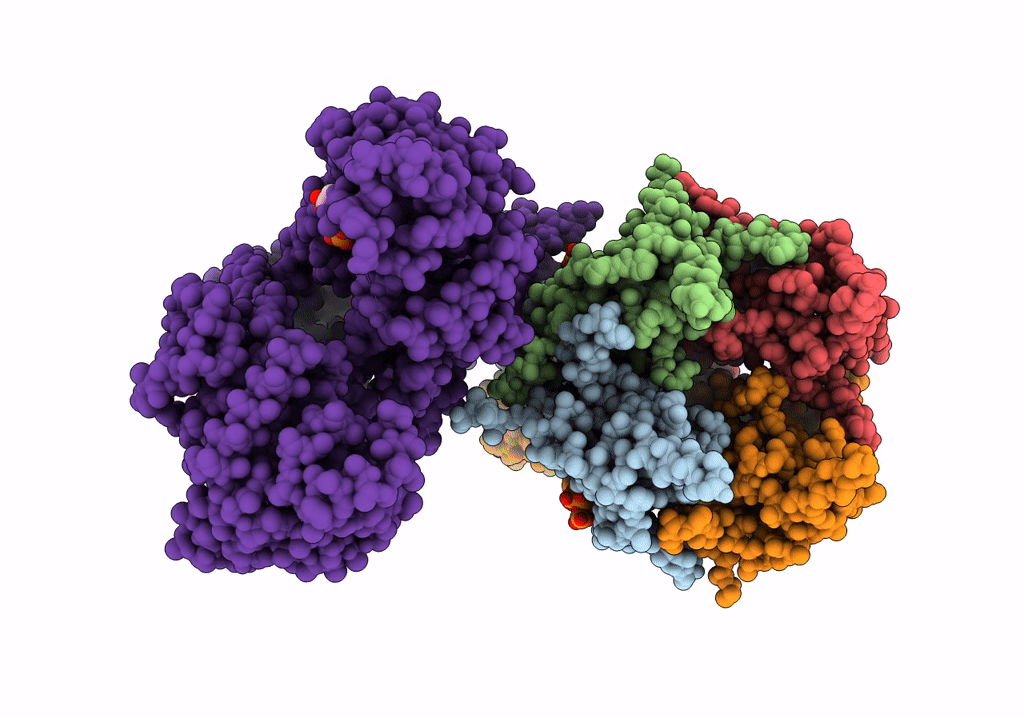
CryoEM structure of the pancreatic ATP-sensitive potassium channel in the ATP-bound state with Kir6.2-CTD in the up conformation
Biological Source:
Source Organism(s):
Rattus norvegicus (Taxon ID: 10116)
Cricetus cricetus (Taxon ID: 10034)
Cricetus cricetus (Taxon ID: 10034)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
5.70 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE