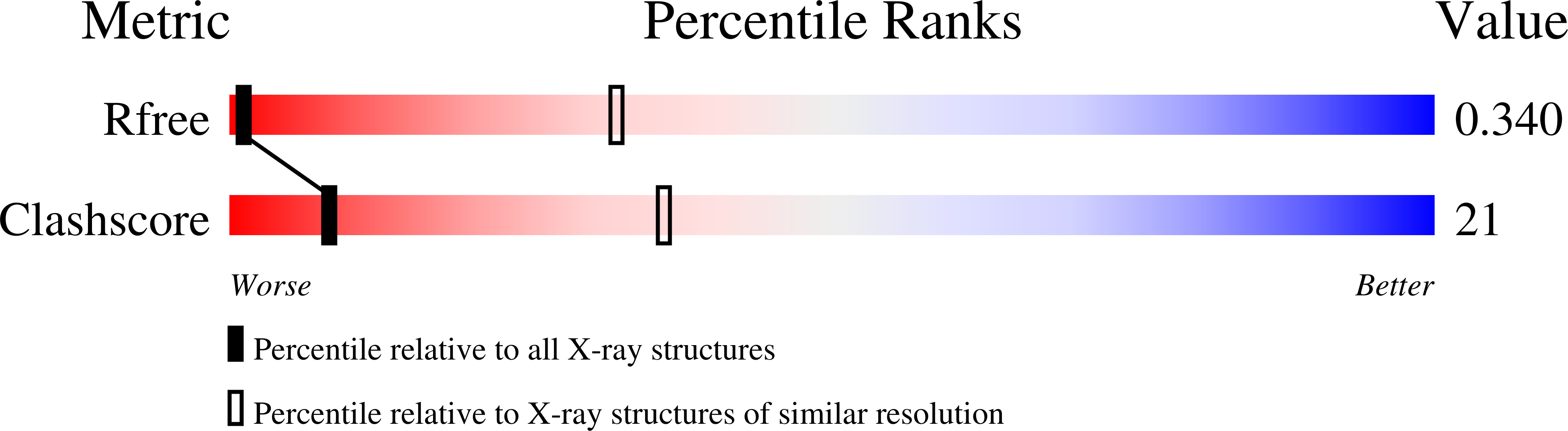
Deposition Date
2022-02-28
Release Date
2022-09-28
Last Version Date
2023-10-25
Entry Detail
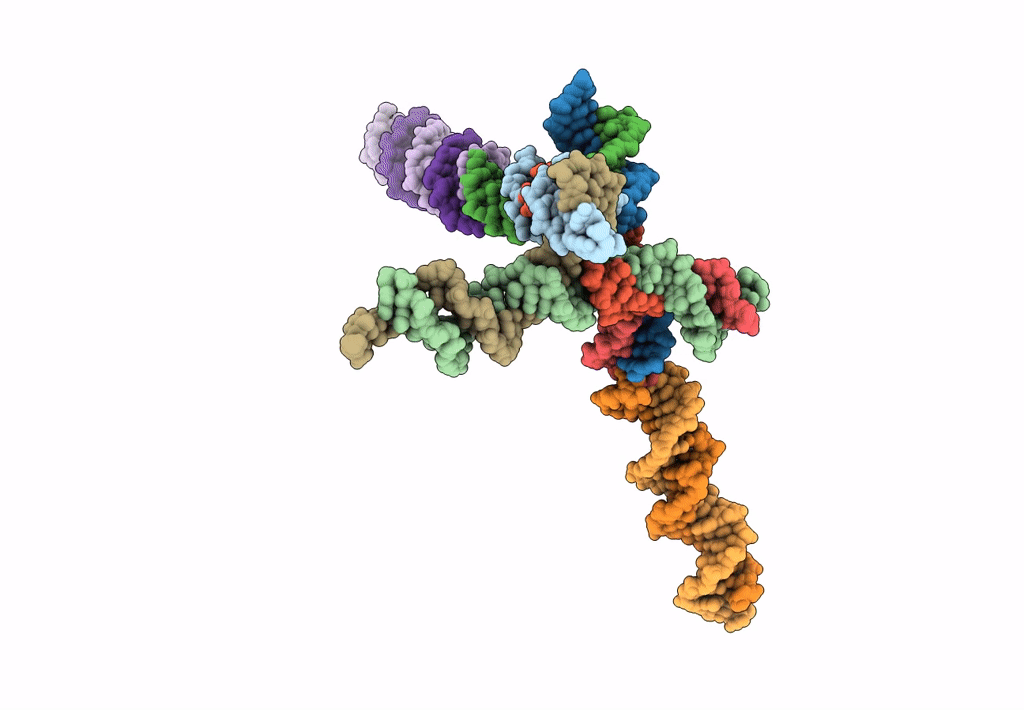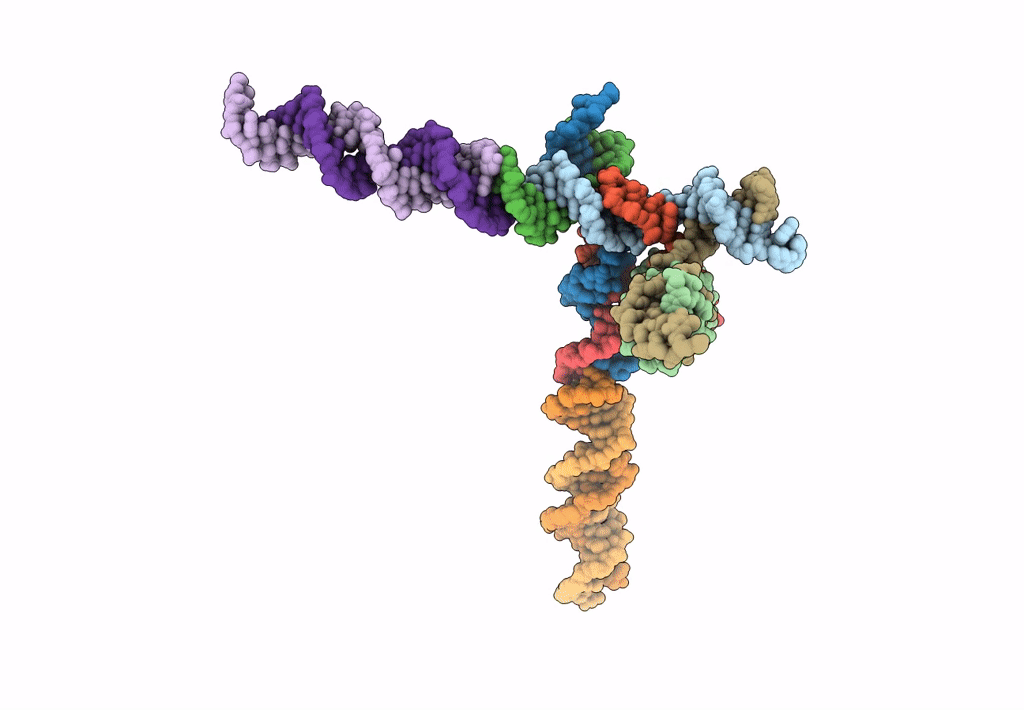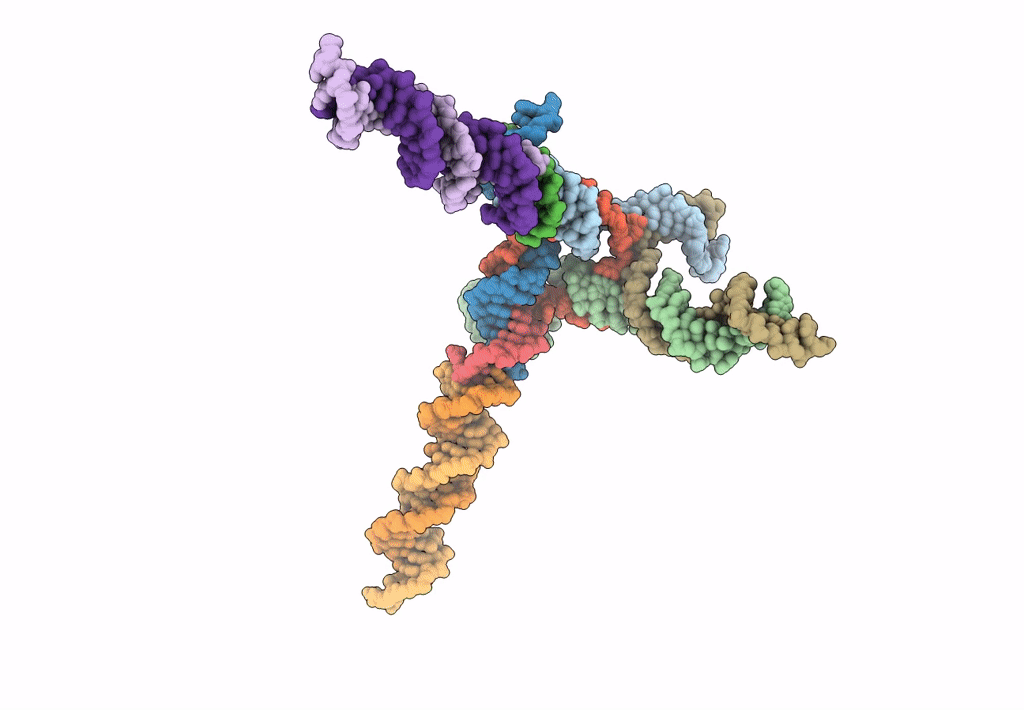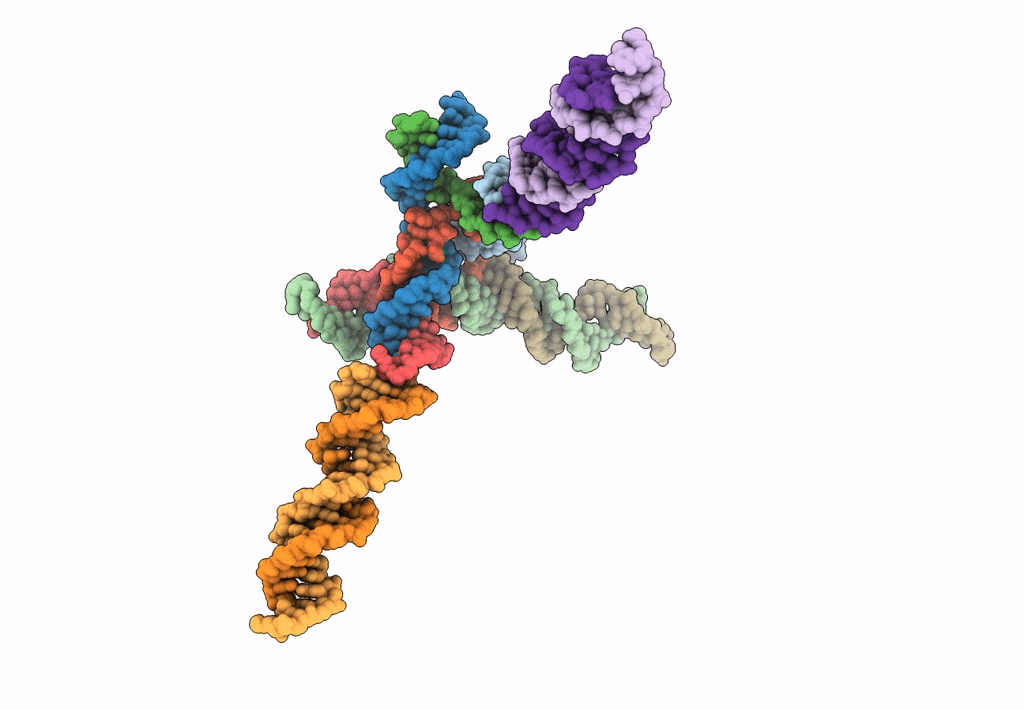
PDB ID:
7U45
Keywords:
Title:
[L344] Self-assembling tensegrity triangle with three turns, four turns and four turns of DNA per axis by extension and linker addition with P1 symmetry
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Resolution:
8.05 Å
R-Value Free:
0.28
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
P 1