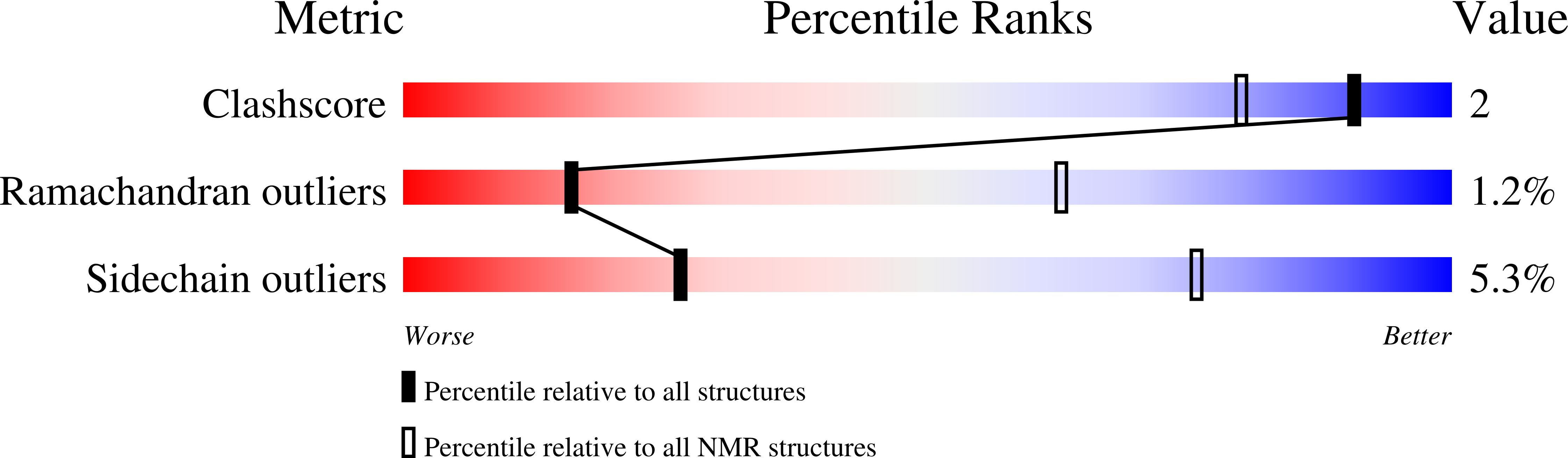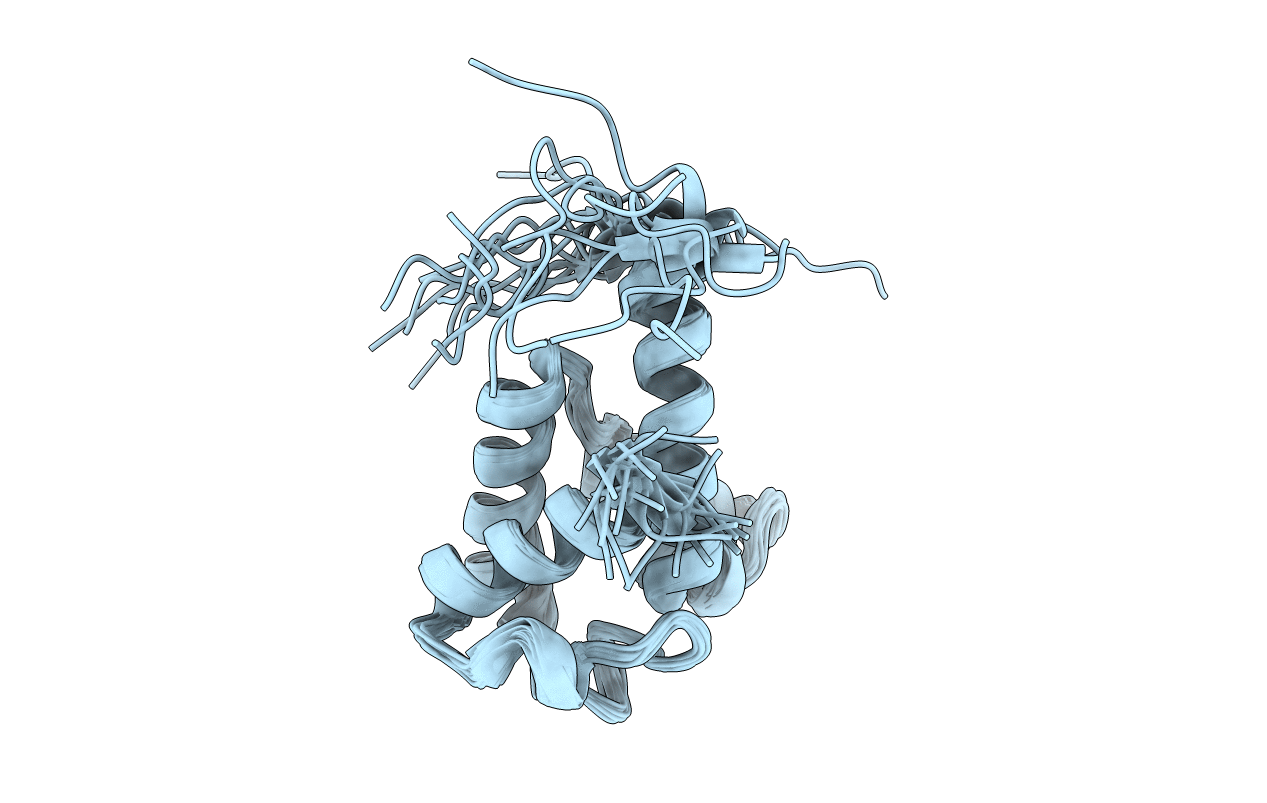
Deposition Date
2022-02-15
Release Date
2022-08-10
Last Version Date
2024-05-15
Entry Detail
PDB ID:
7TZD
Keywords:
Title:
Solution NMR of the specialized apo-acyl carrier protein (RPA2022) from Rhodopseudomonas palustris, refined without RDCs. Northeast Structural Genomics Consortium Target RpR324
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
100
Conformers Submitted:
20
Selection Criteria:
structures with the lowest energy