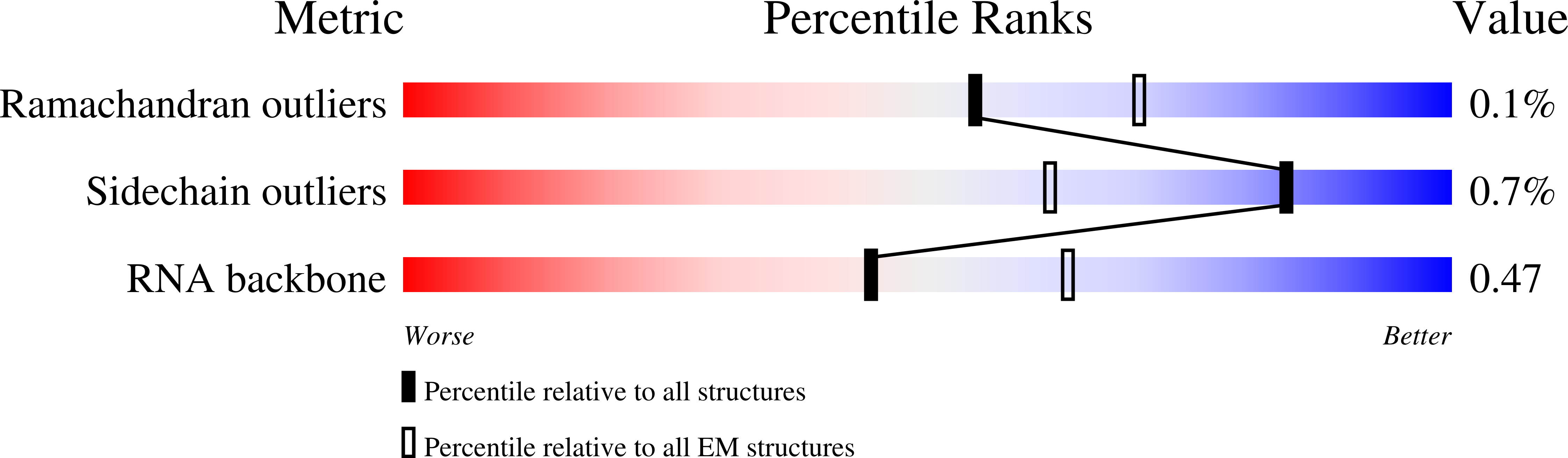
Deposition Date
2022-02-03
Release Date
2022-10-19
Last Version Date
2024-06-12
Entry Detail
PDB ID:
7TUT
Keywords:
Title:
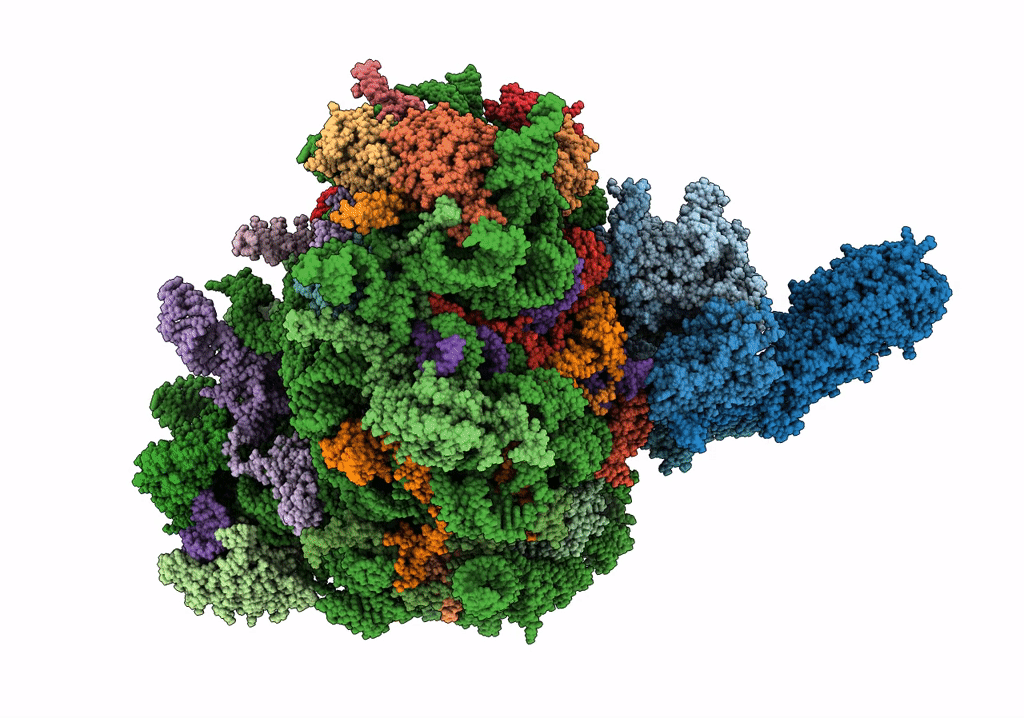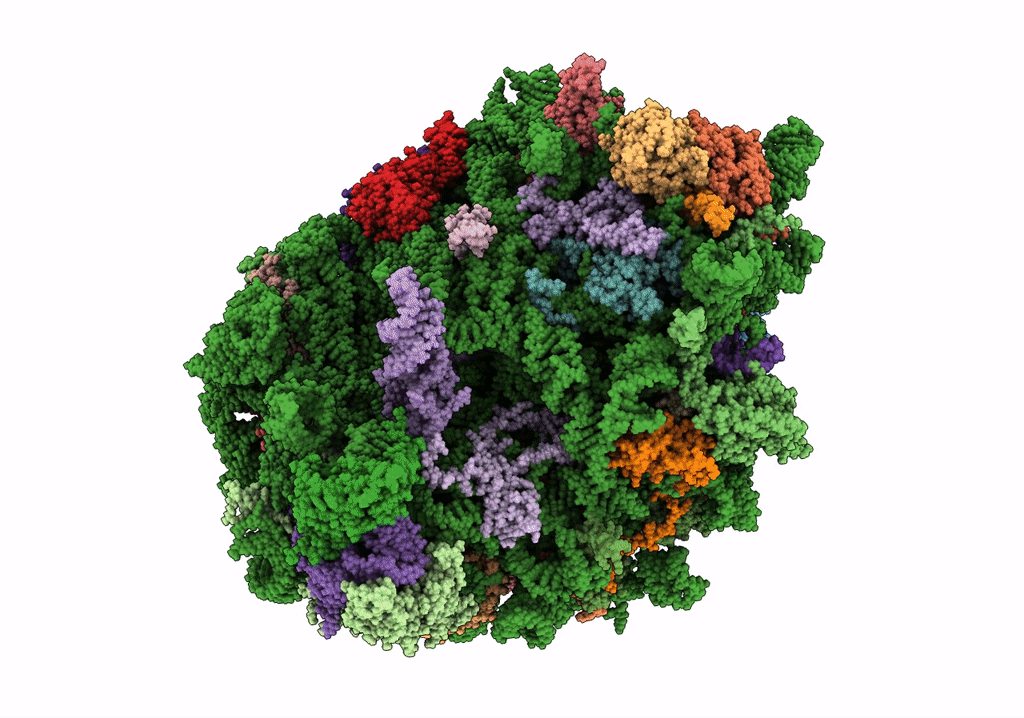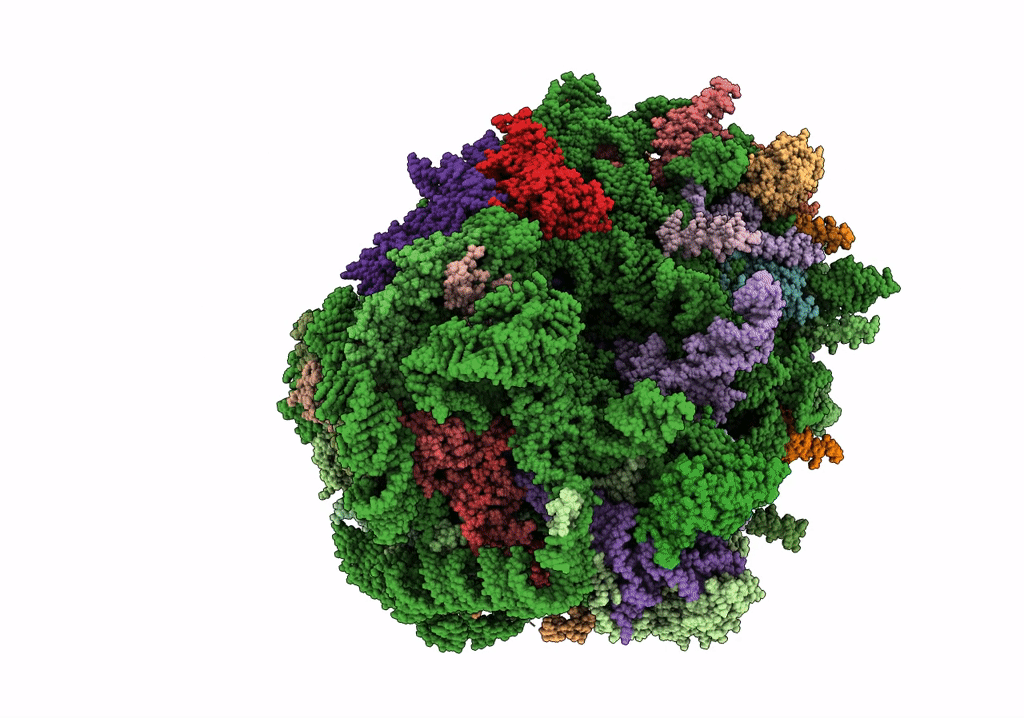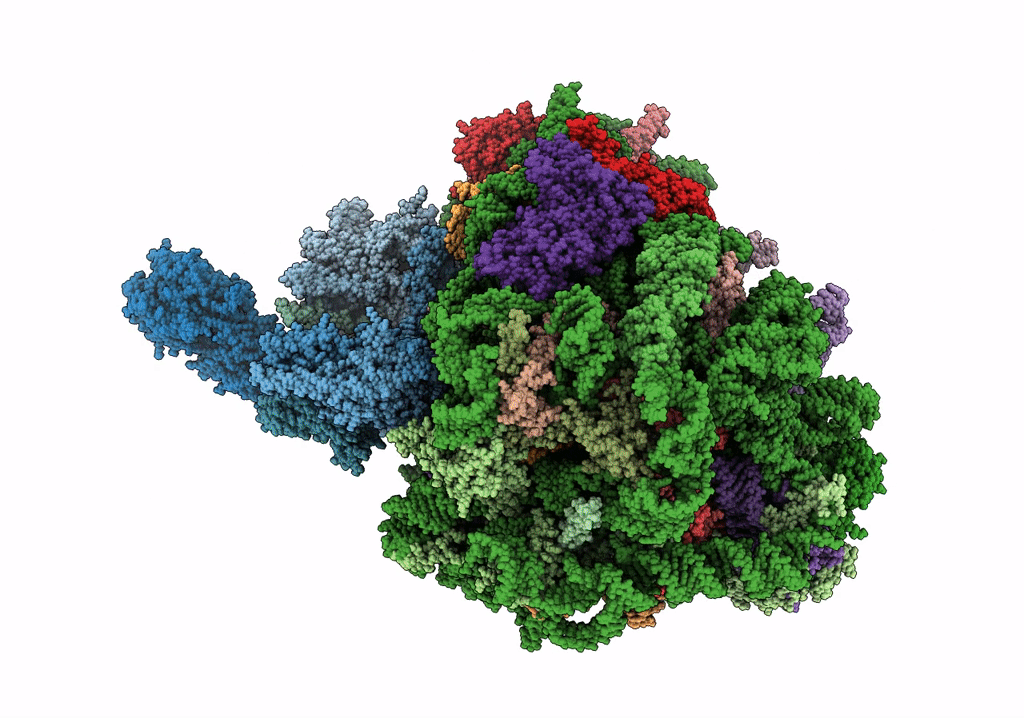
Structure of the rabbit 80S ribosome stalled on a 4-TMD Rhodopsin intermediate in complex with the multipass translocon
Biological Source:
Source Organism(s):
Oryctolagus cuniculus (Taxon ID: 9986)
Canis lupus (Taxon ID: 9612)
Canis lupus (Taxon ID: 9612)
Method Details:
Experimental Method:
Resolution:
3.88 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE