Deposition Date
2022-01-25
Release Date
2023-01-04
Last Version Date
2023-10-25
Entry Detail
PDB ID:
7TP6
Keywords:
Title:
The crystal structure of T252E CYP199A4 bound to 4-methylthiobenzoic acid
Biological Source:
Source Organism:
Rhodopseudomonas palustris HaA2 (Taxon ID: 316058)
Host Organism:
Method Details:
Experimental Method:
Resolution:
1.66 Å
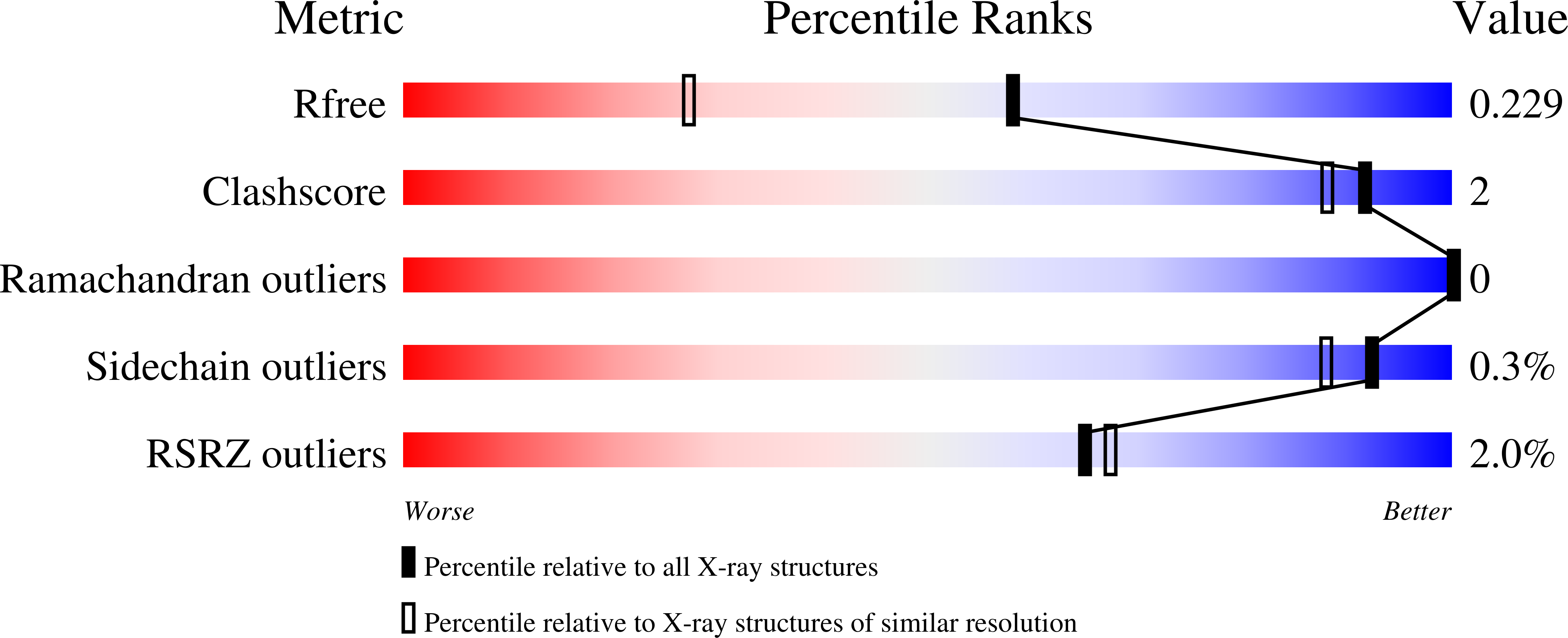
R-Value Free:
0.23
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 1 21 1