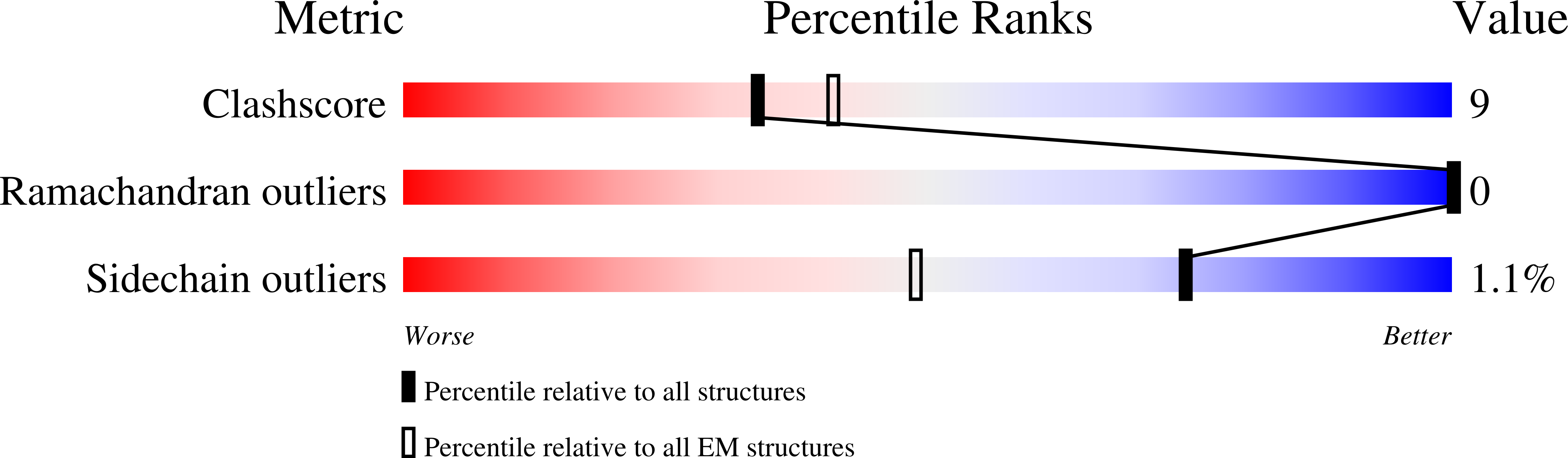
Deposition Date
2022-01-13
Release Date
2022-02-16
Last Version Date
2024-02-28
Entry Detail
PDB ID:
7TID
Keywords:
Title:
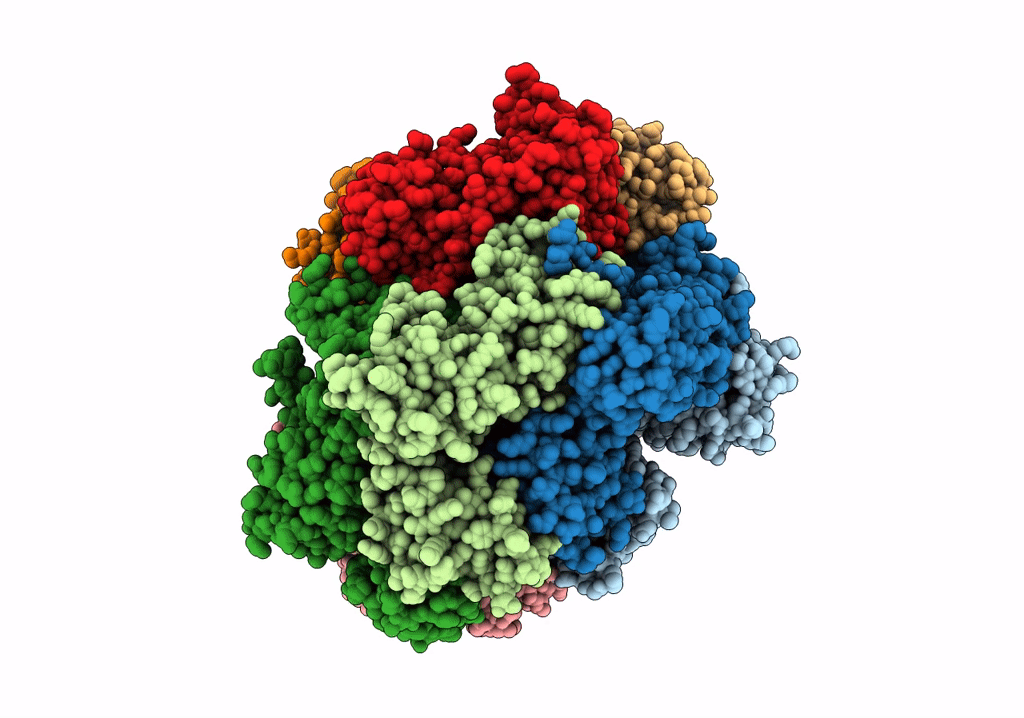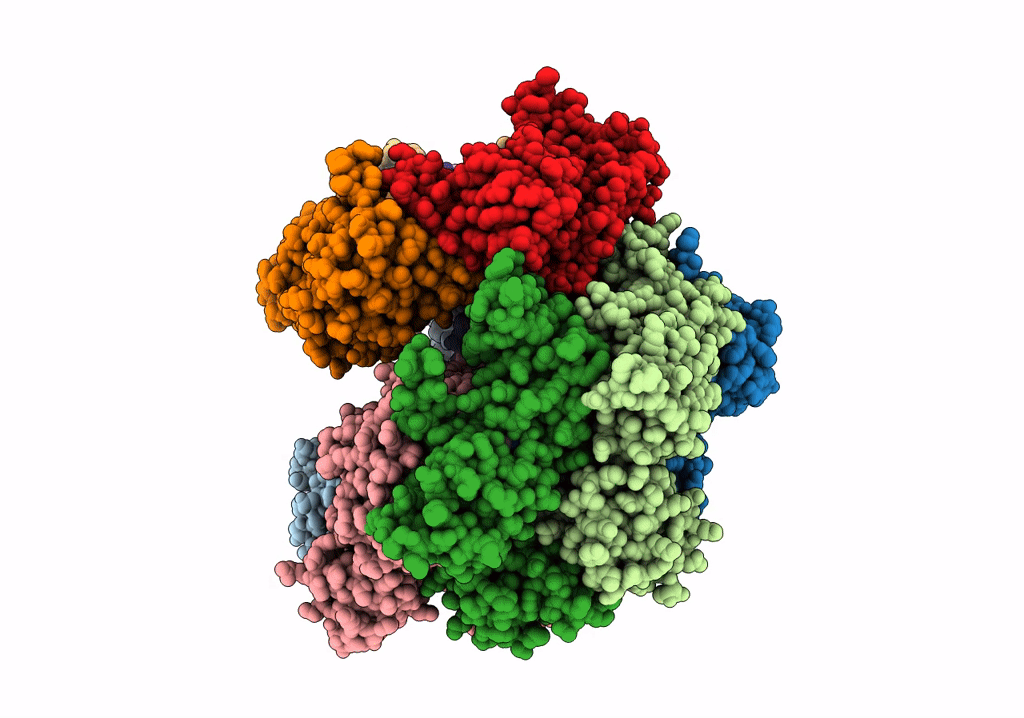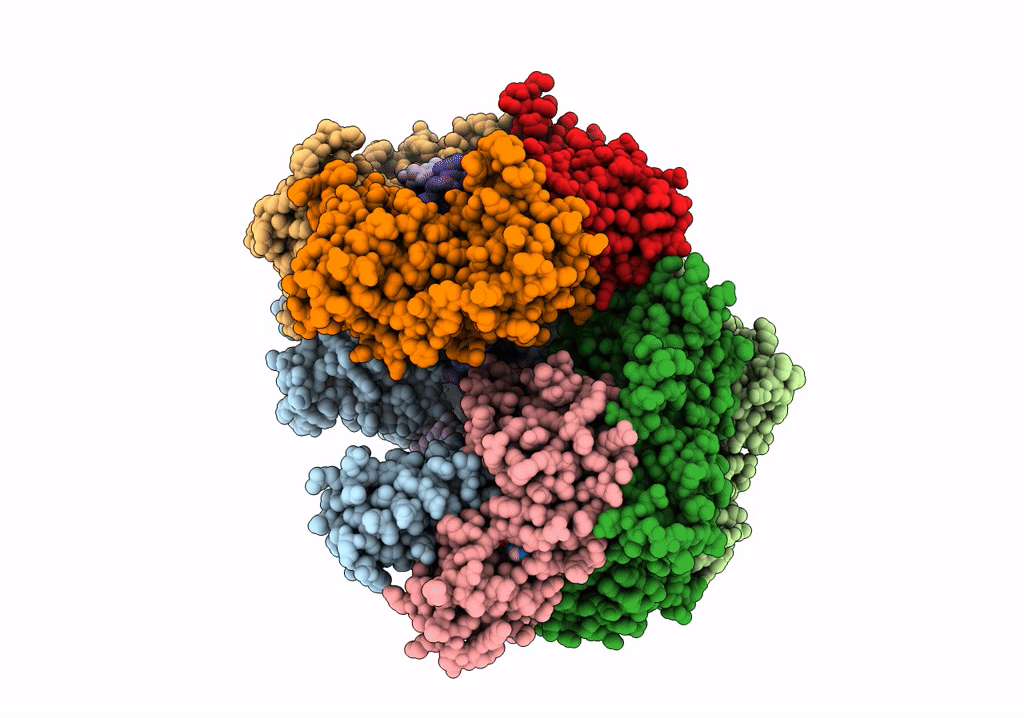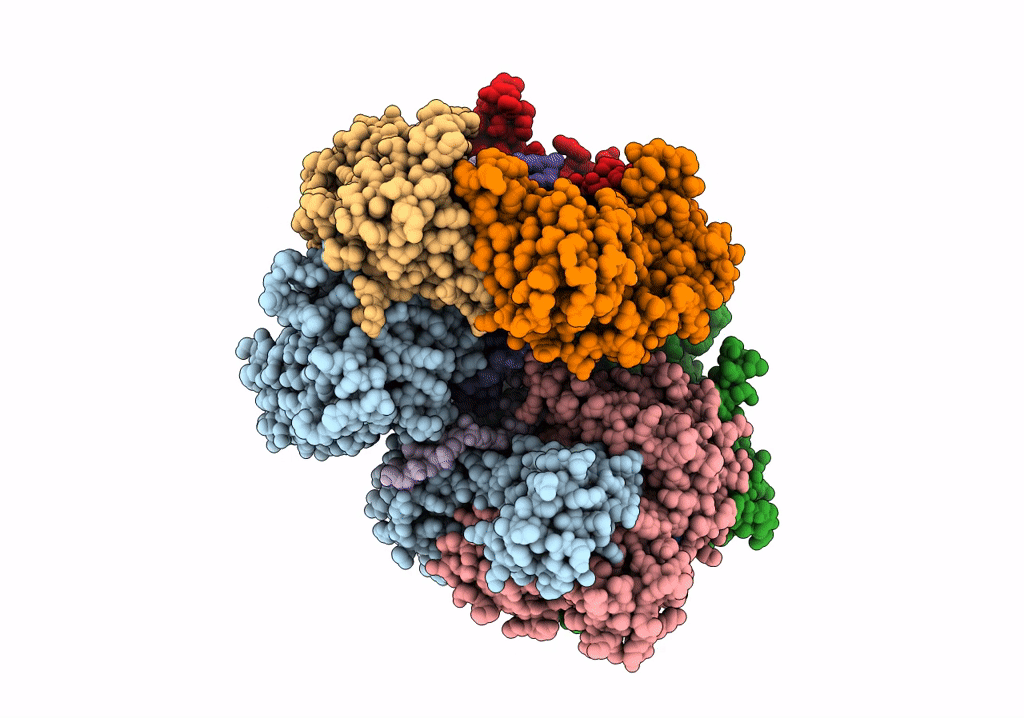
Structure of the yeast clamp loader (Replication Factor C RFC) bound to the sliding clamp (Proliferating Cell Nuclear Antigen PCNA) and primer-template DNA
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (Taxon ID: 4932)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE