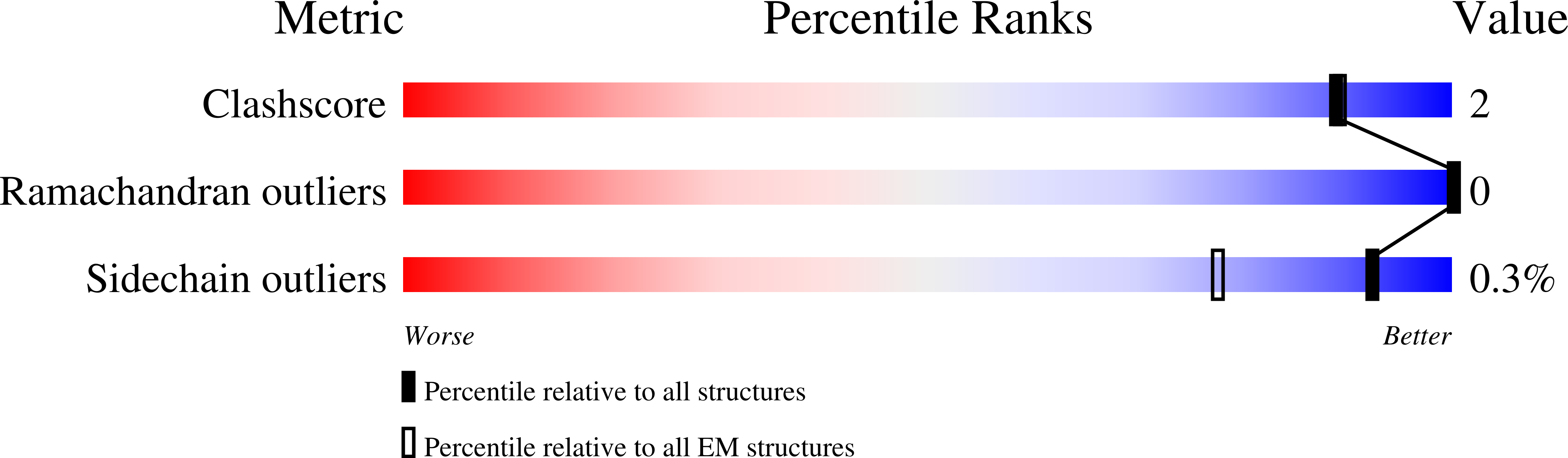
Deposition Date
2021-12-13
Release Date
2023-06-14
Last Version Date
2024-10-23
Entry Detail
PDB ID:
7T5Q
Keywords:
Title:
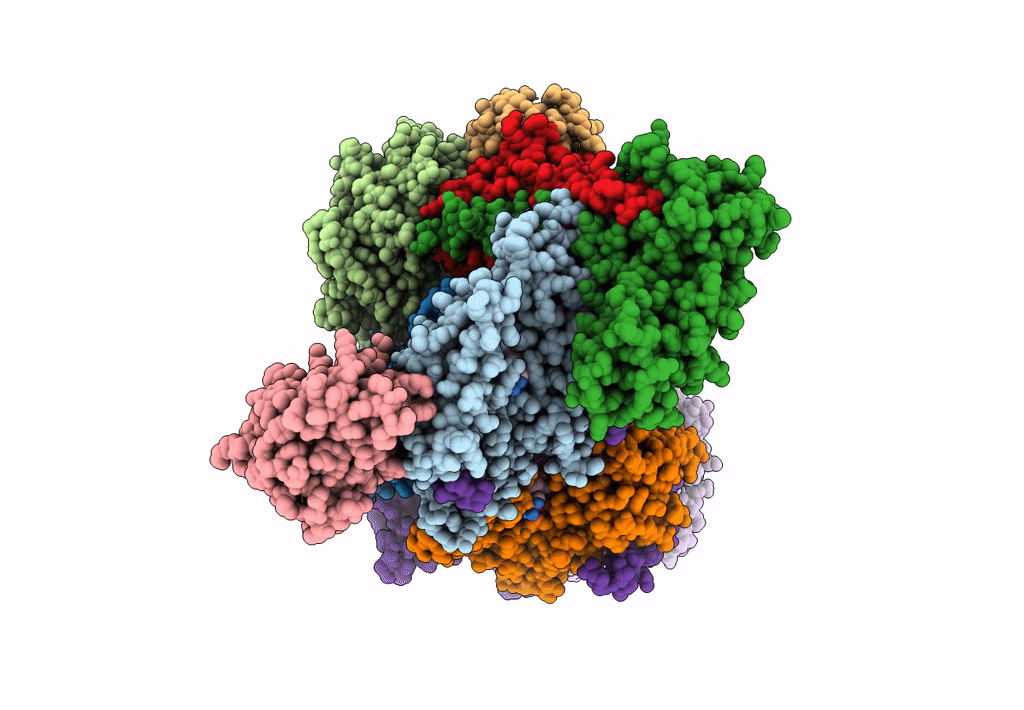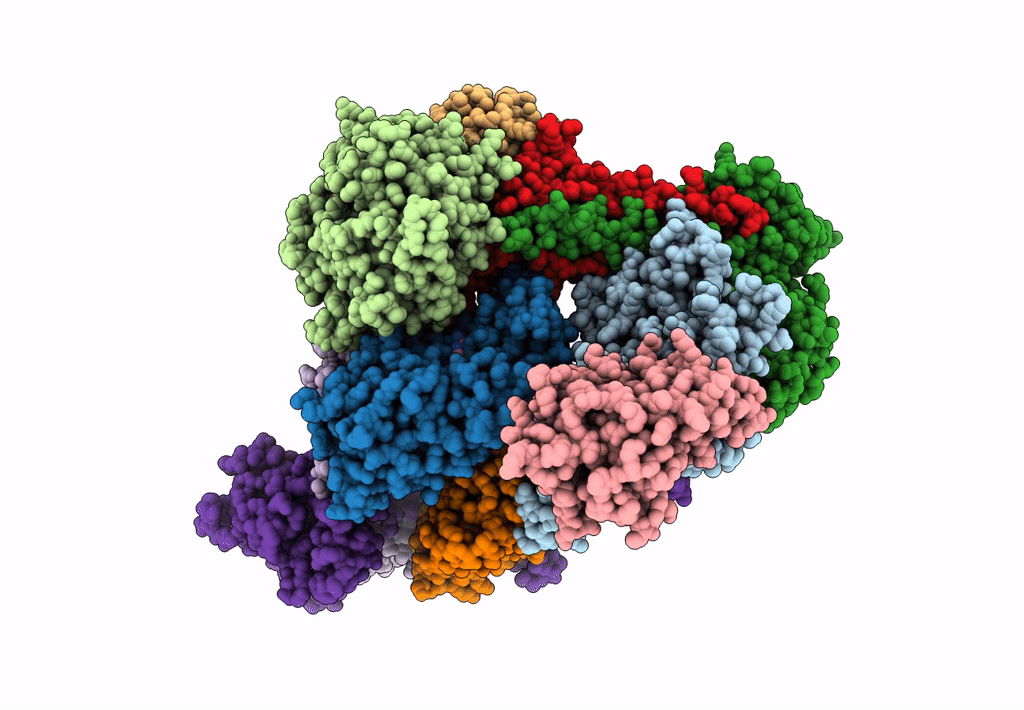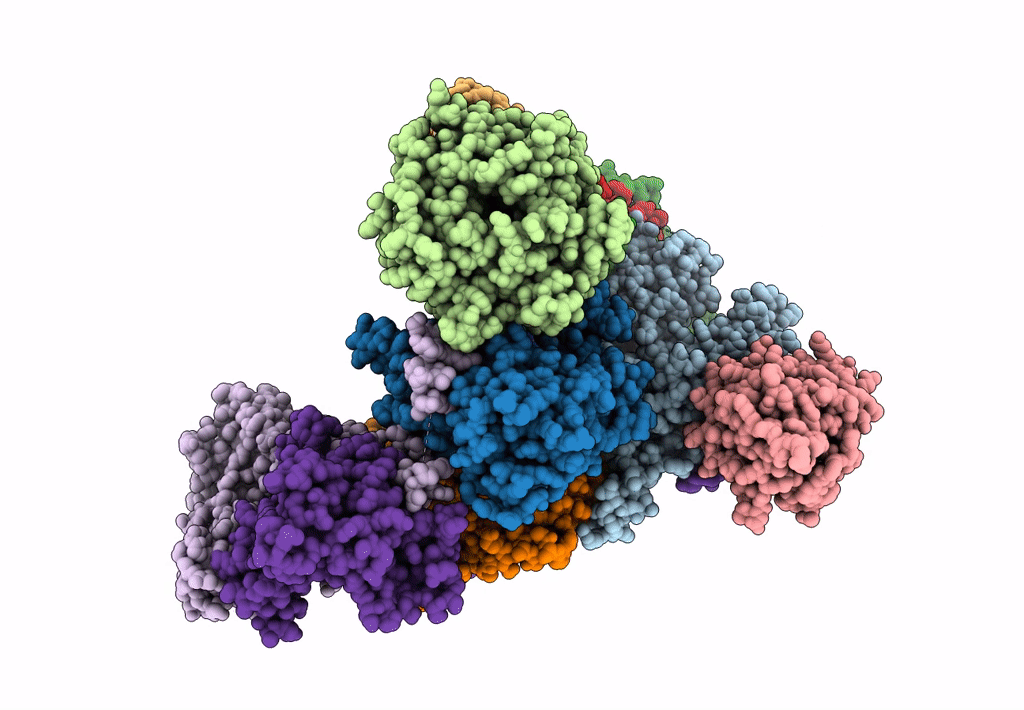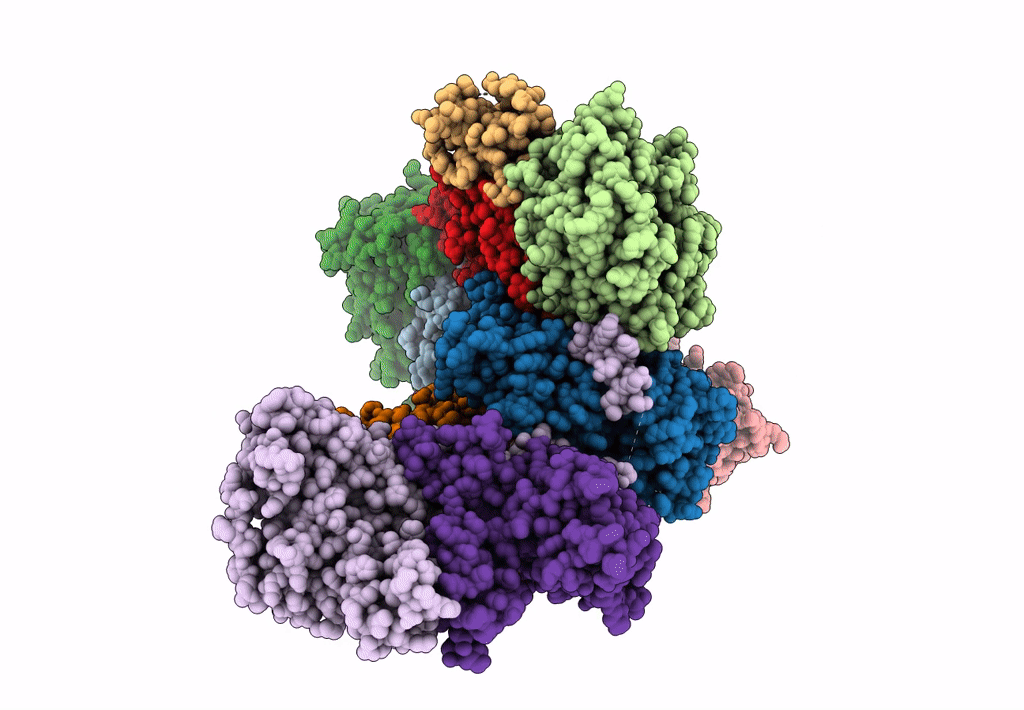
Cryo-EM Structure of a Transition State of Arp2/3 Complex Activation
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Mus musculus (Taxon ID: 10090)
Bos taurus (Taxon ID: 9913)
Oryctolagus cuniculus (Taxon ID: 9986)
Mus musculus (Taxon ID: 10090)
Bos taurus (Taxon ID: 9913)
Oryctolagus cuniculus (Taxon ID: 9986)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.40 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE