Deposition Date
2021-12-06
Release Date
2022-03-16
Last Version Date
2024-12-25
Entry Detail
PDB ID:
7T30
Keywords:
Title:
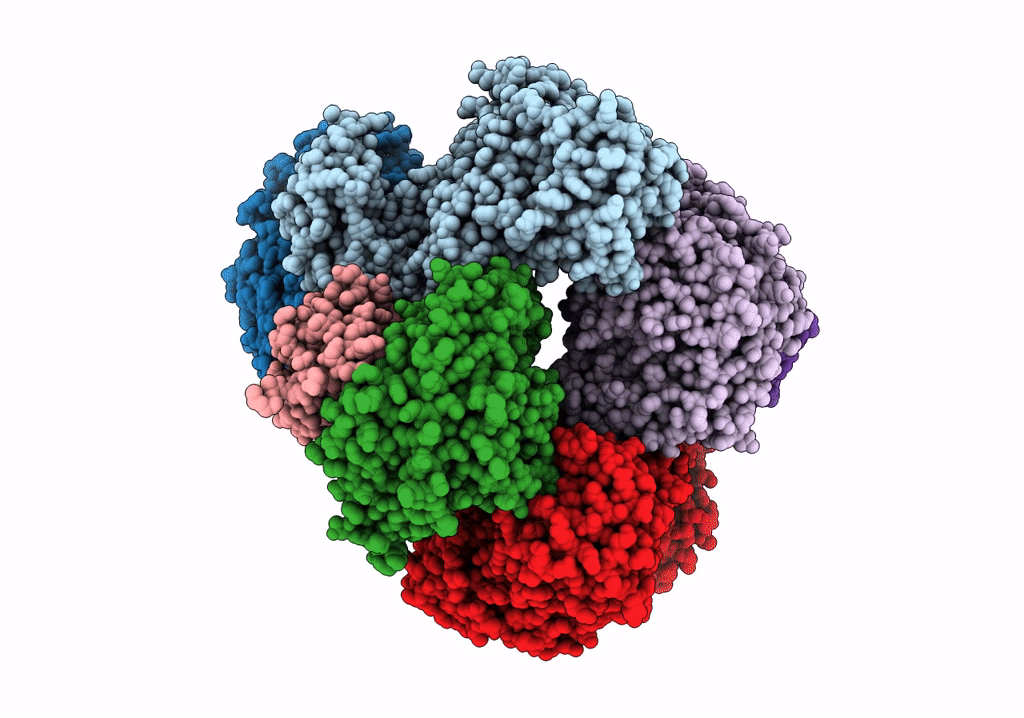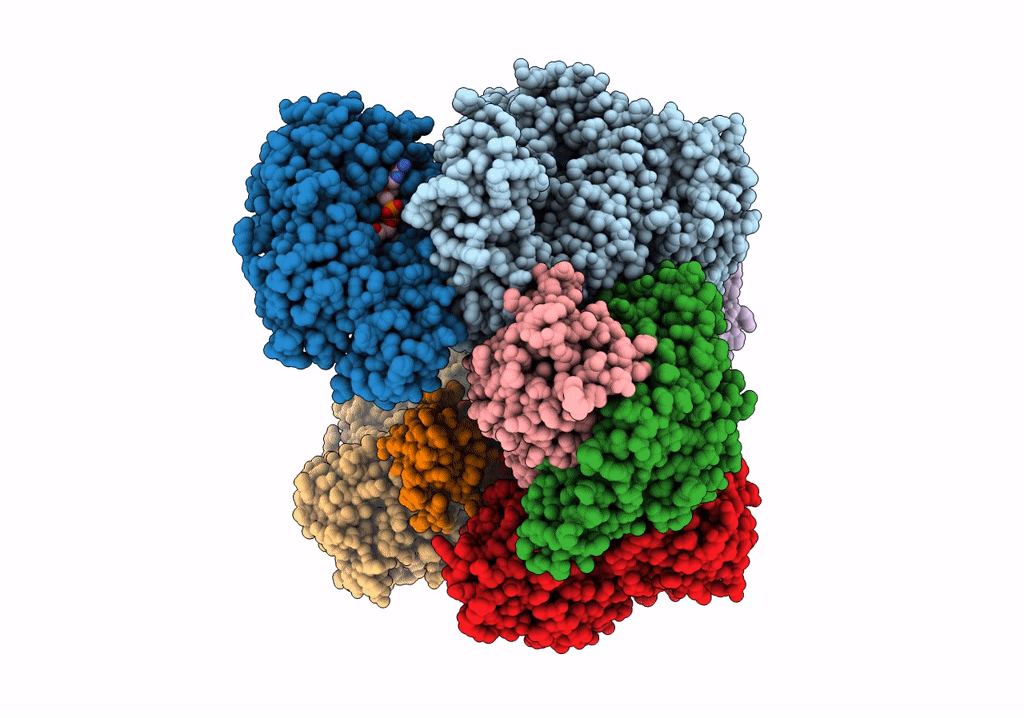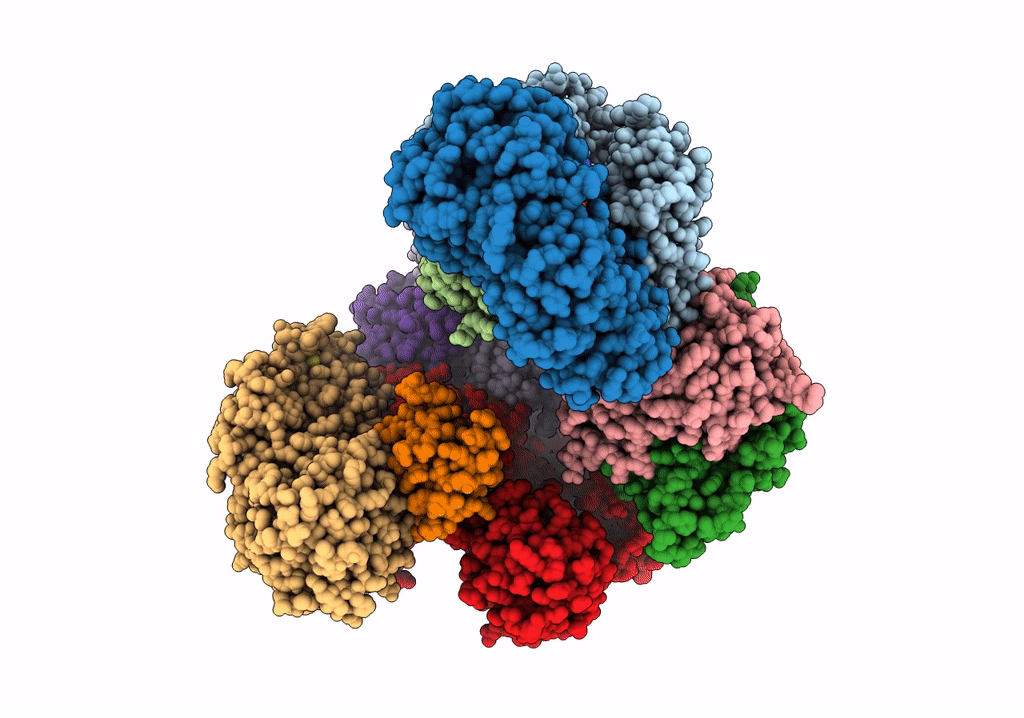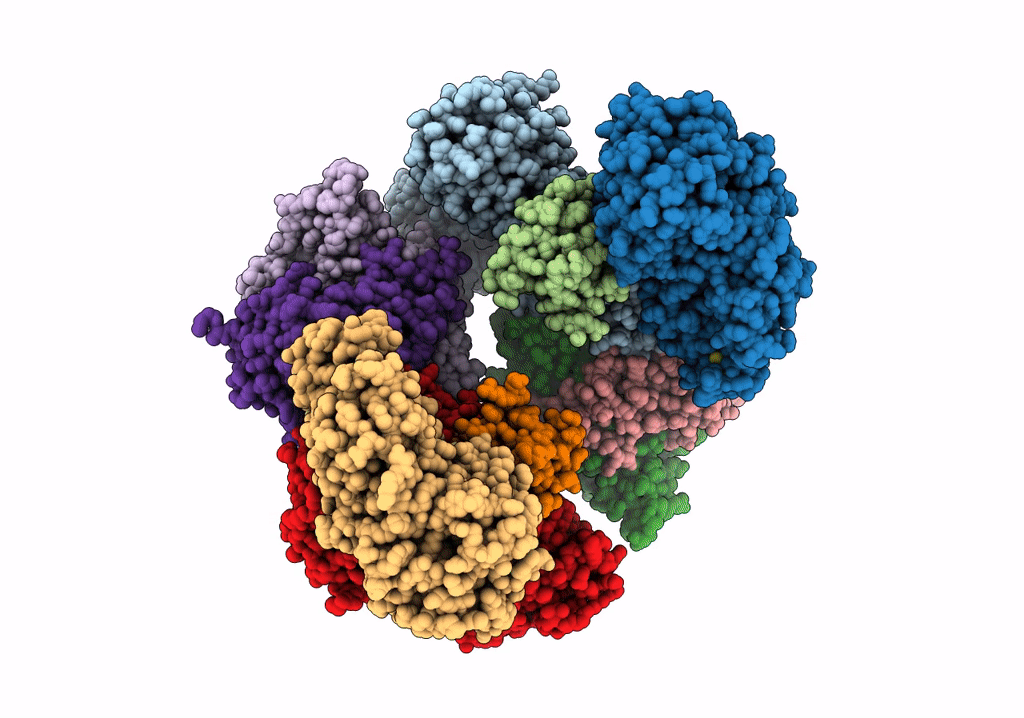
Structure of electron bifurcating Ni-Fe hydrogenase complex HydABCSL in FMN/NAD(H) bound state
Biological Source:
Source Organism(s):
Acetomicrobium mobile (Taxon ID: 97477)
Method Details:
Experimental Method:
Resolution:
3.00 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


