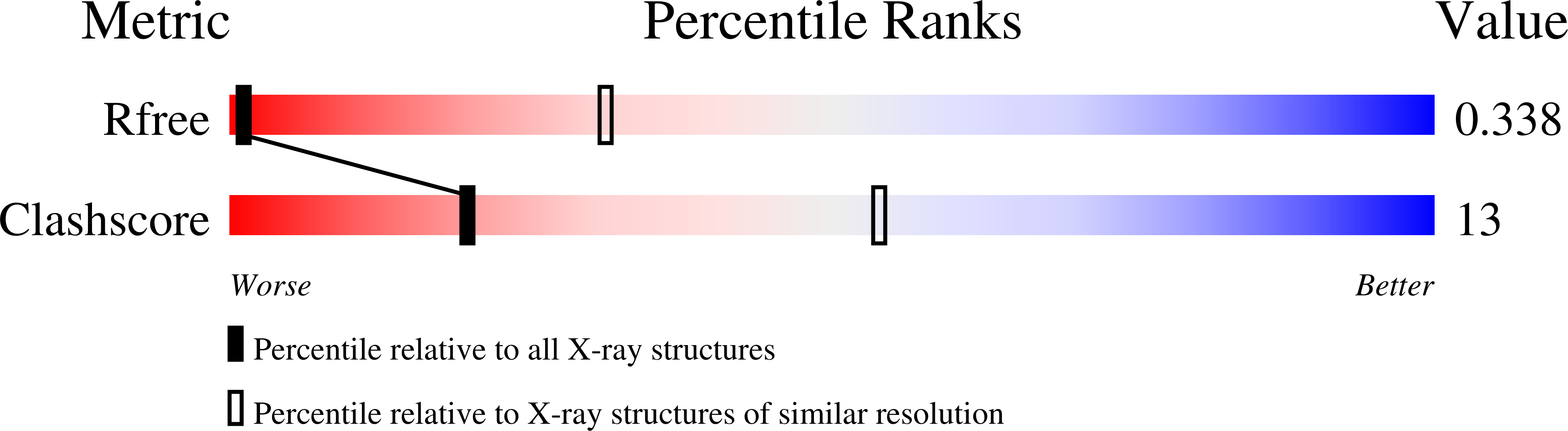Deposition Date
2021-11-02
Release Date
2022-11-09
Last Version Date
2023-10-25
Entry Detail
PDB ID:
7SPL
Keywords:
Title:
[2T3] Self-assembling 3D DNA triangle with three inter-junction base pairs containing the L1 junction and a zero-linked center strand
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Method Details:
Experimental Method:
Resolution:
6.09 Å
R-Value Free:
0.33
R-Value Work:
0.23
R-Value Observed:
0.23
Space Group:
H 3