Deposition Date
2022-01-14
Release Date
2022-10-12
Last Version Date
2024-01-31
Entry Detail
PDB ID:
7QT1
Keywords:
Title:
Non-obligately L8S8-complex forming RubisCO derived from ancestral sequence reconstruction and rational engineering in L8S8 complex with substitution e170N
Biological Source:
Source Organism(s):
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
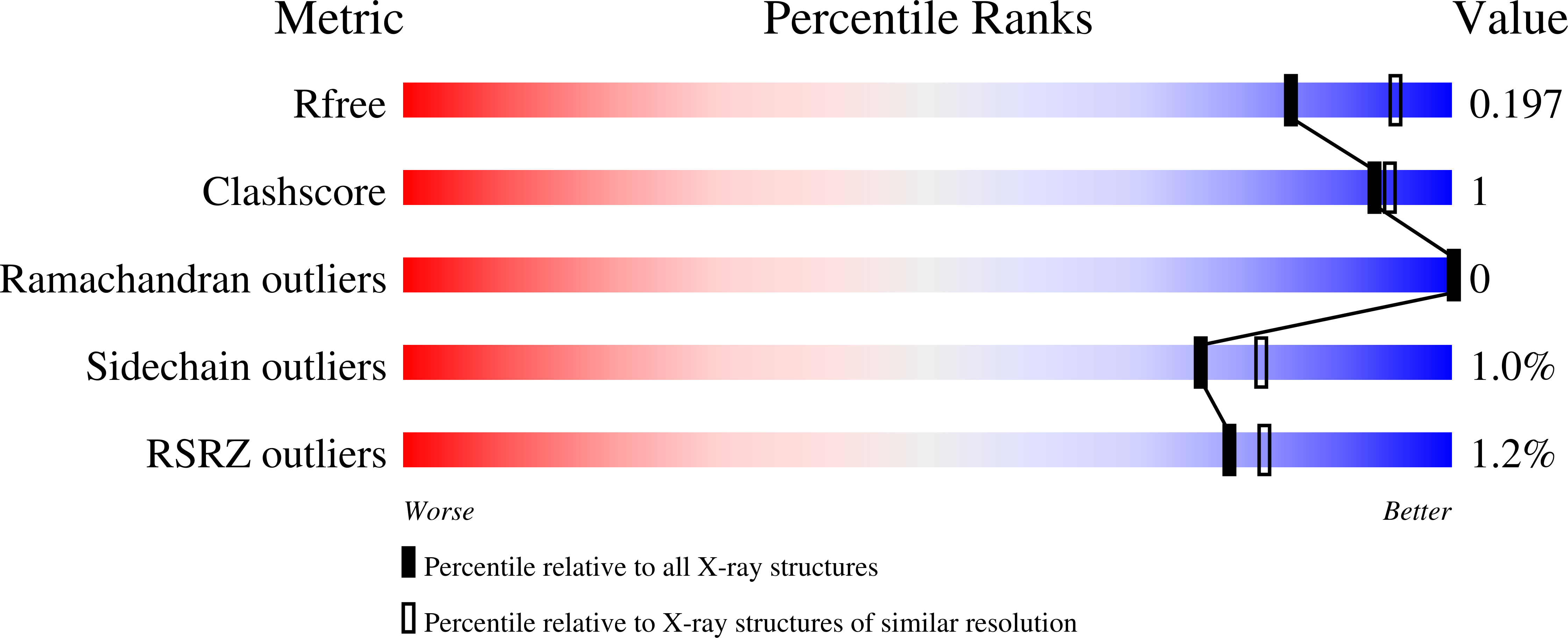
Resolution:
2.10 Å
R-Value Free:
0.18
R-Value Work:
0.16
R-Value Observed:
0.16
Space Group:
I 1 2 1