Deposition Date
2021-12-08
Release Date
2022-09-28
Last Version Date
2024-01-31
Entry Detail
PDB ID:
7QGC
Keywords:
Title:
H. SAPIENS CK2 KINASE ALPHA SUBUNIT IN COMPLEX WITH THE ATP-COMPETITIVE INHIBITOR 5,6-DIBROMOBENZOTRIAZOLE AT PH 5.5
Biological Source:
Source Organism(s):
Homo sapiens (Taxon ID: 9606)
Expression System(s):
Method Details:
Experimental Method:
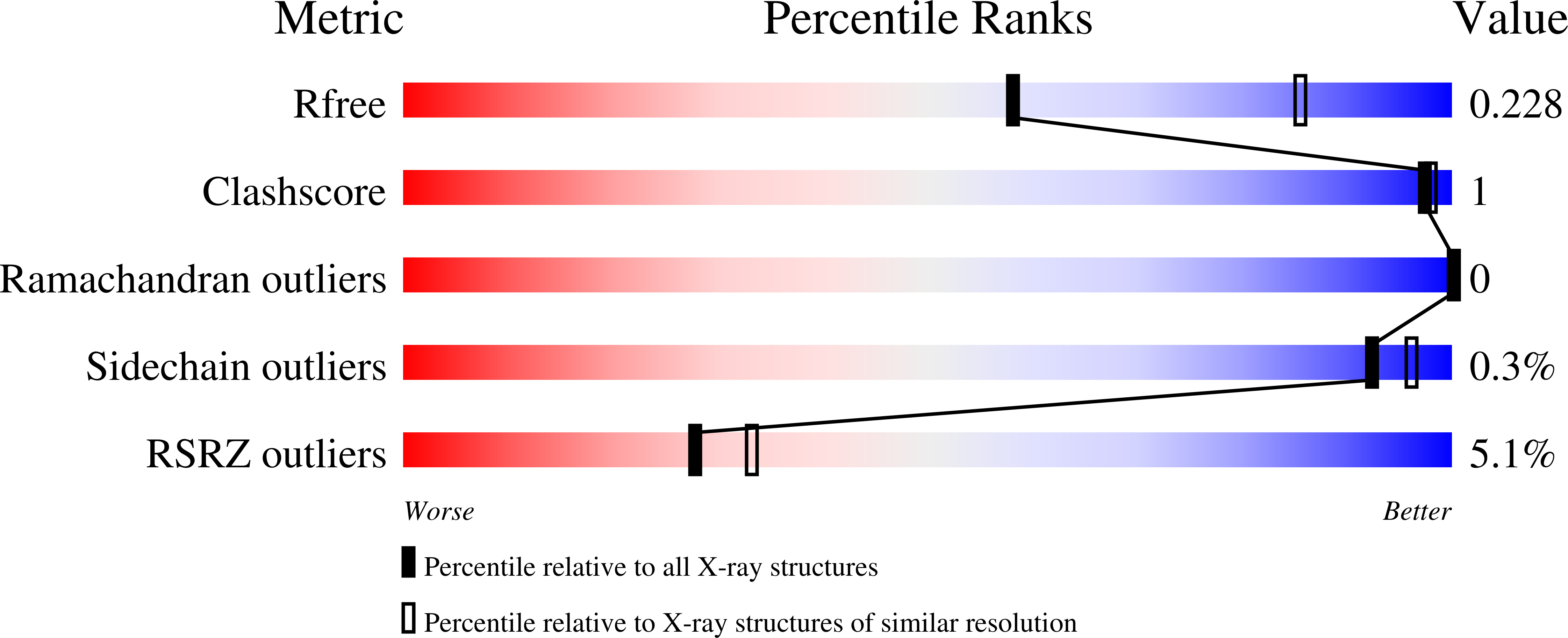
Resolution:
2.55 Å
R-Value Free:
0.23
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 42 21 2