Deposition Date
2021-11-26
Release Date
2022-10-26
Last Version Date
2024-01-10
Entry Detail
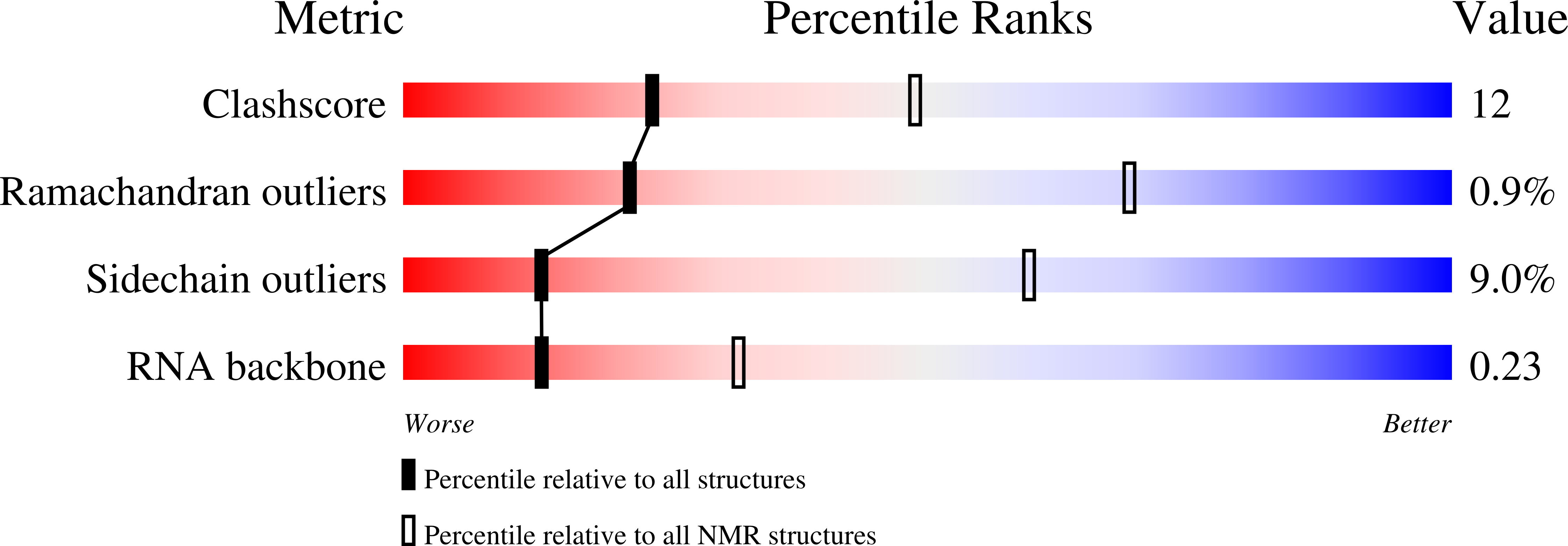
PDB ID:
7QDD
Keywords:
Title:
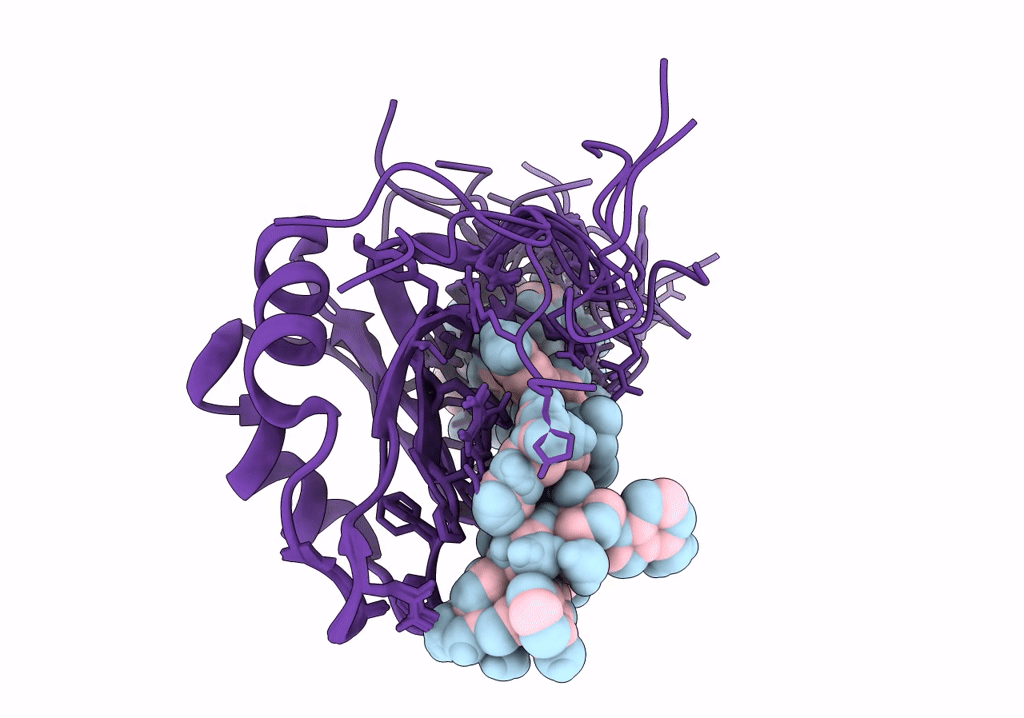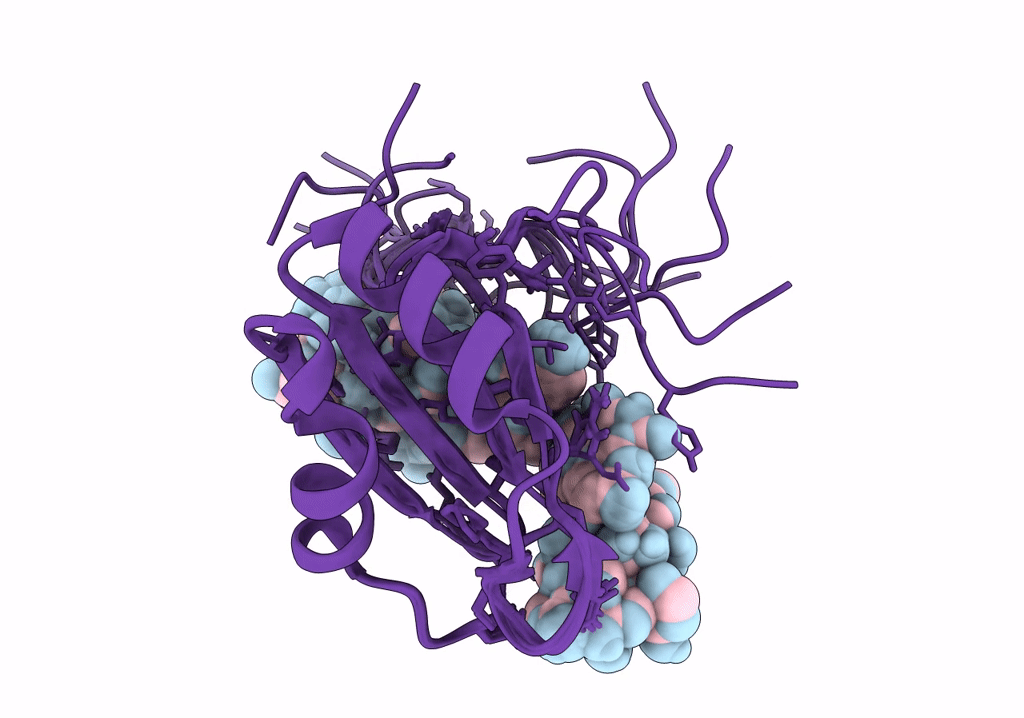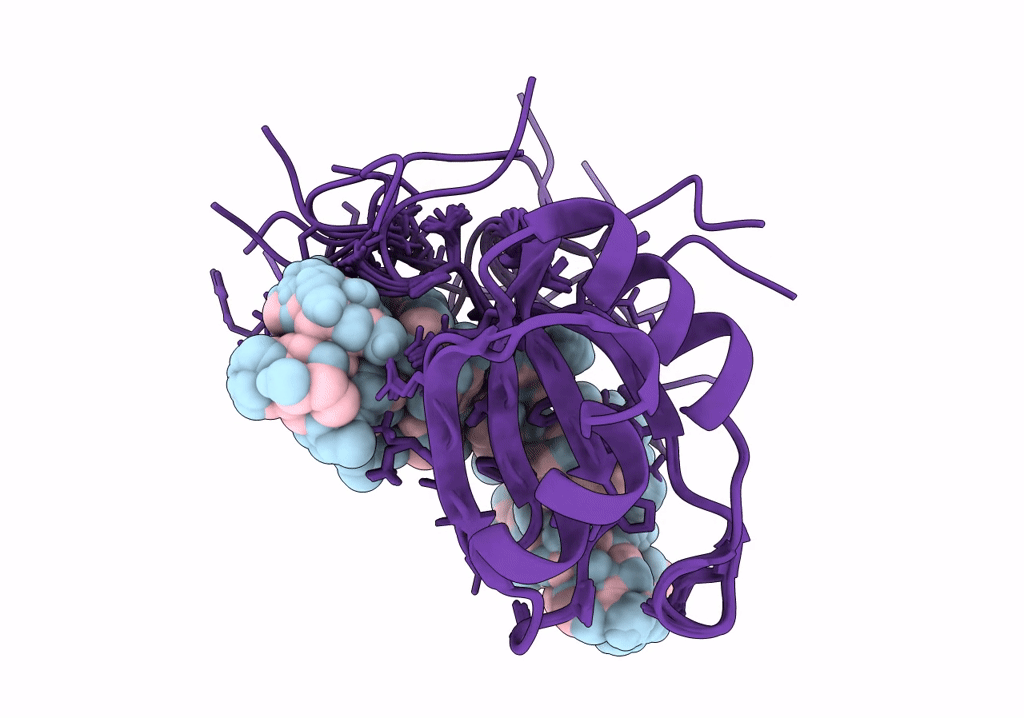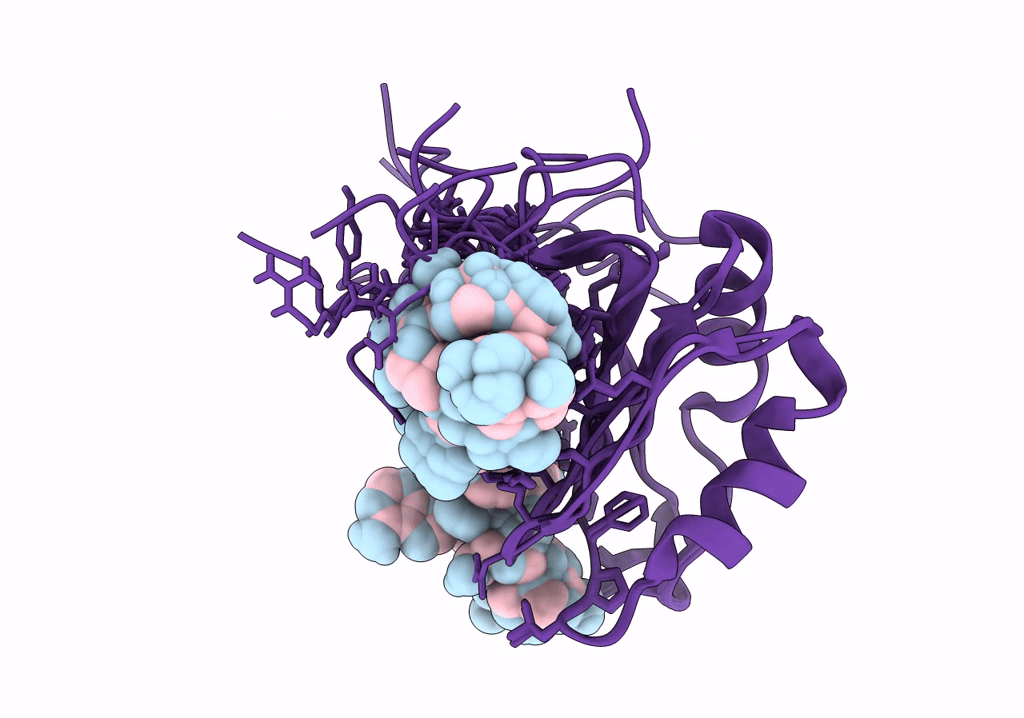
NMR structure of Npl3 RRM1 bound to the AUCCAA RNA
Biological Source:
Source Organism(s):
Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Taxon ID: 559292)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Conformers Calculated:
50
Conformers Submitted:
10
Selection Criteria:
structures with the lowest energy