Deposition Date
2021-11-03
Release Date
2022-11-16
Last Version Date
2024-07-17
Entry Detail
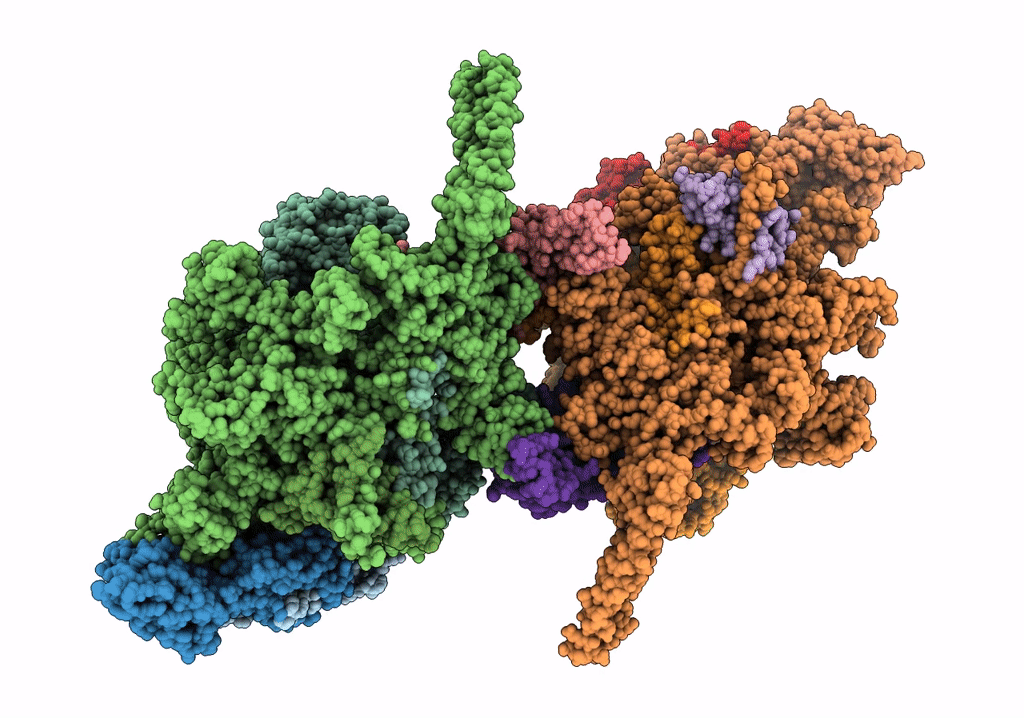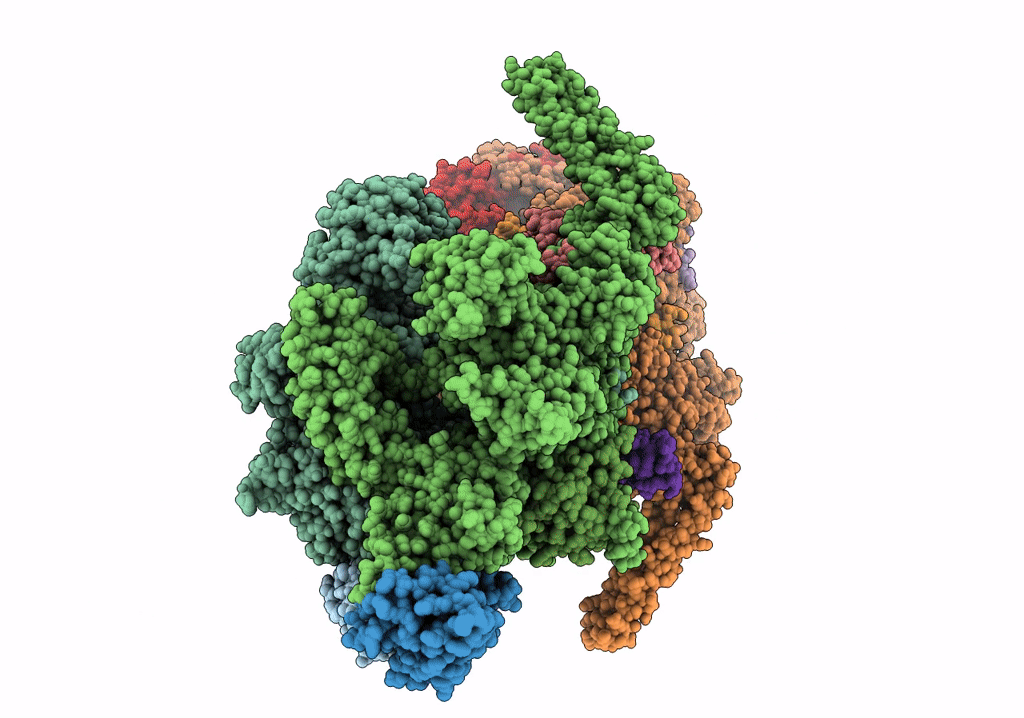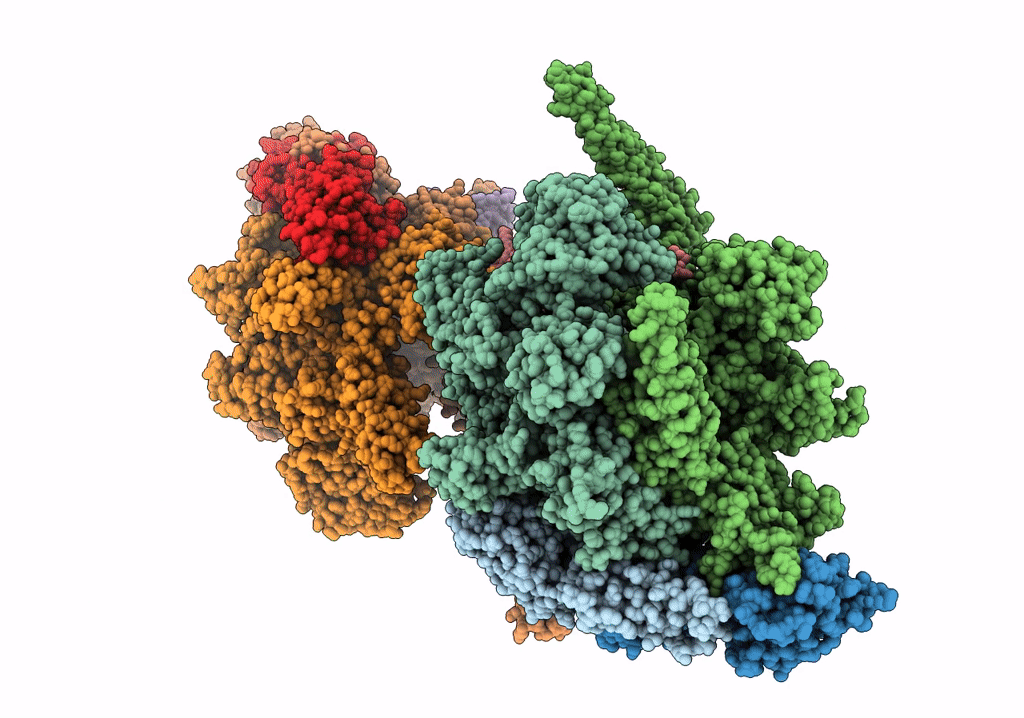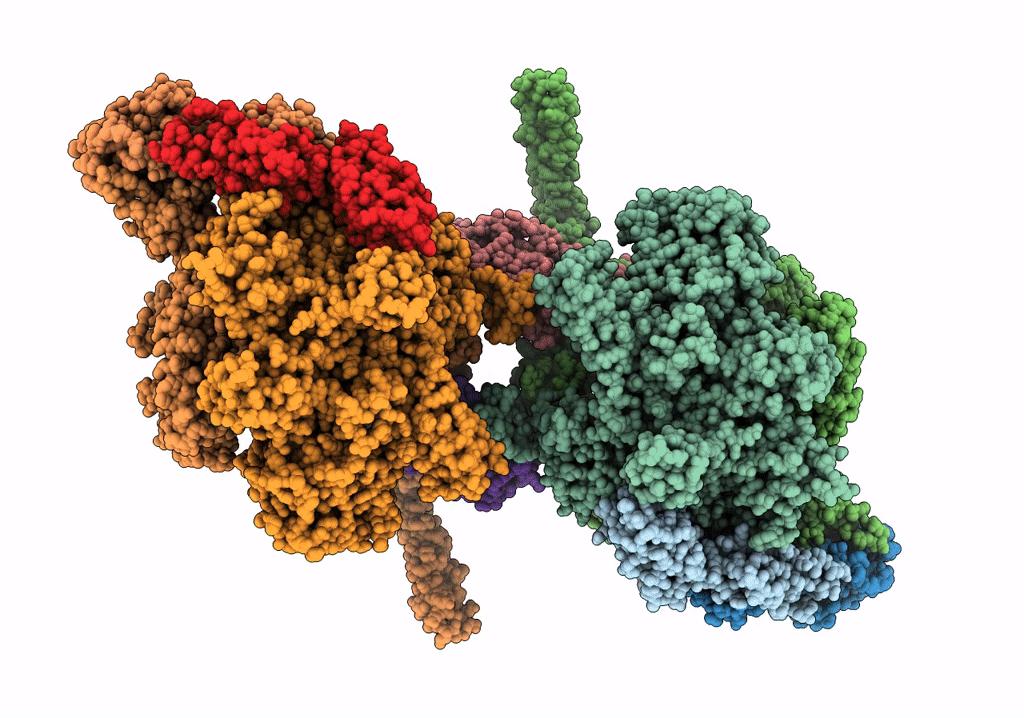
PDB ID:
7Q59
Keywords:
Title:
Cryo-EM structure of Mycobacterium tuberculosis RNA polymerase holoenzyme dimer comprising sigma factor SigB
Biological Source:
Source Organism(s):
Mycobacterium tuberculosis H37Rv (Taxon ID: 83332)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
4.36 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE