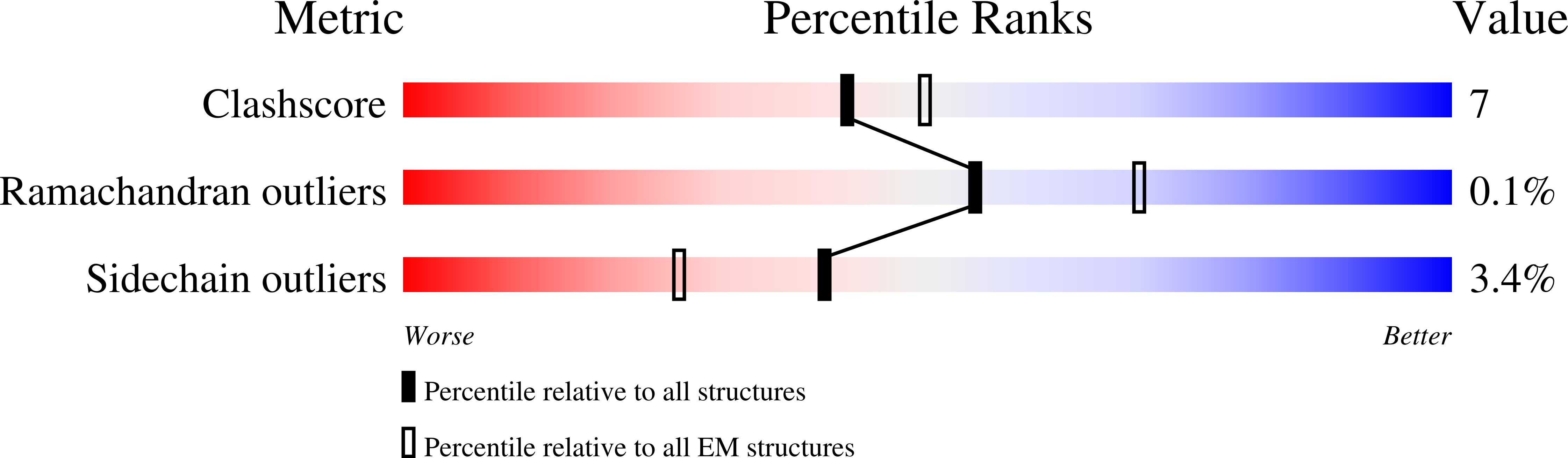
Deposition Date
2021-10-08
Release Date
2022-01-19
Last Version Date
2024-07-17
Entry Detail
PDB ID:
7PXC
Keywords:
Title:
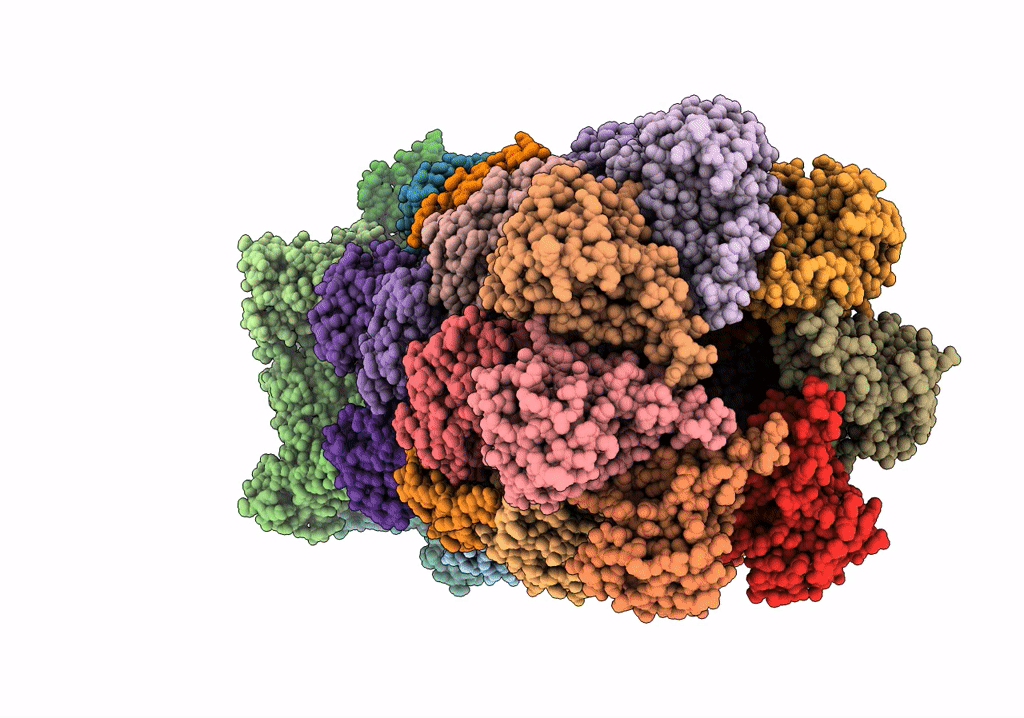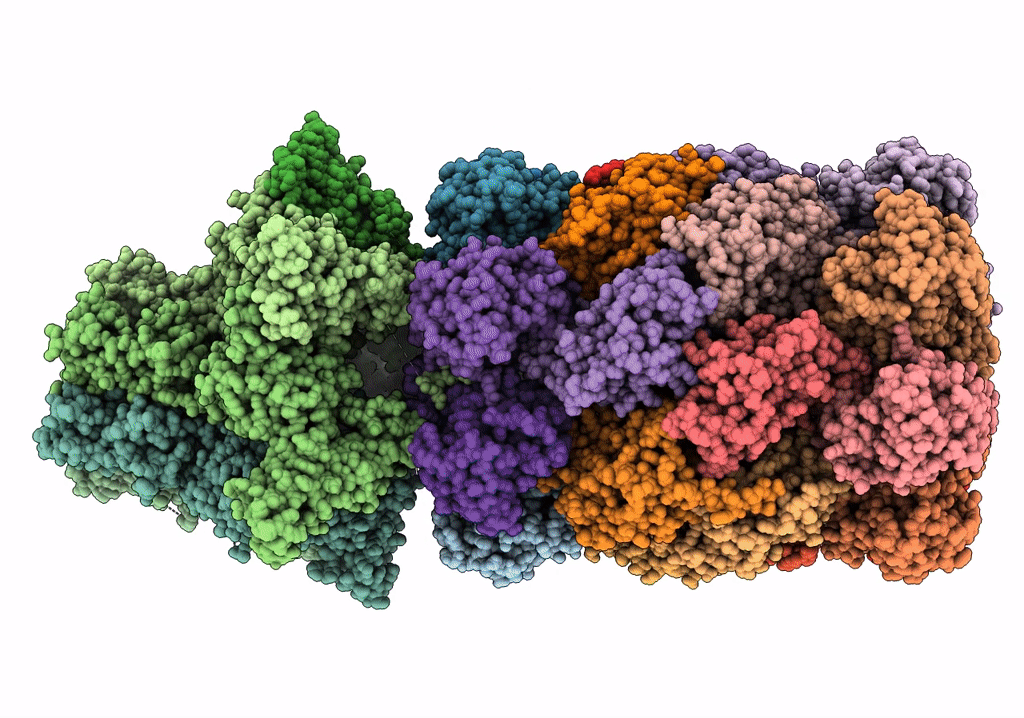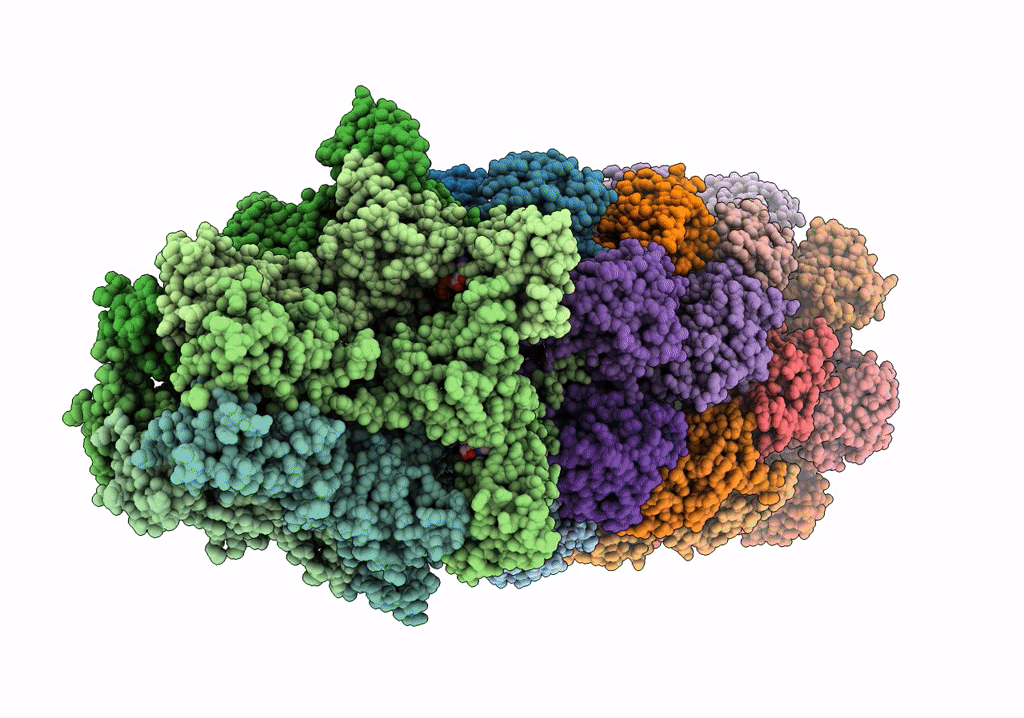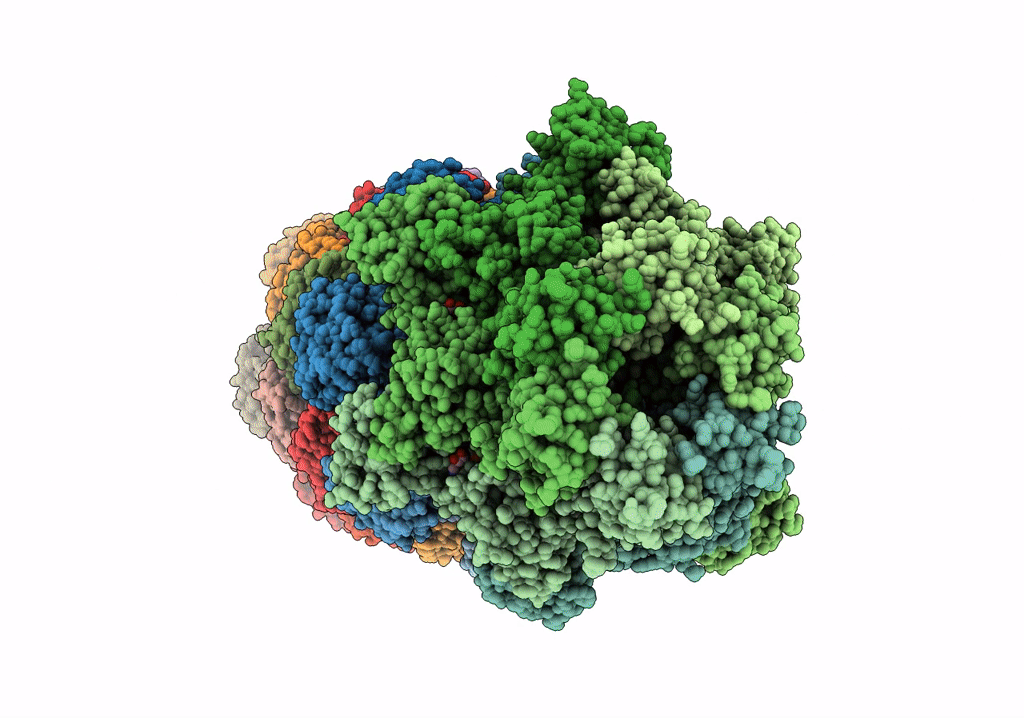
Substrate-engaged mycobacterial Proteasome-associated ATPase in complex with open-gate 20S CP - composite map (state A)
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.84 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE