Deposition Date
2021-09-13
Release Date
2022-07-06
Last Version Date
2024-01-31
Entry Detail
PDB ID:
7PP3
Keywords:
Title:
STRUCTURE OF ESTER-HYDROLASE EH7 FROM THE METAGENOME OF MARINE SEDIMENTS AT MILAZZO HARBOR (SICILY, ITALY)
Biological Source:
Source Organism(s):
uncultured bacterium (Taxon ID: 77133)
Expression System(s):
Method Details:
Experimental Method:
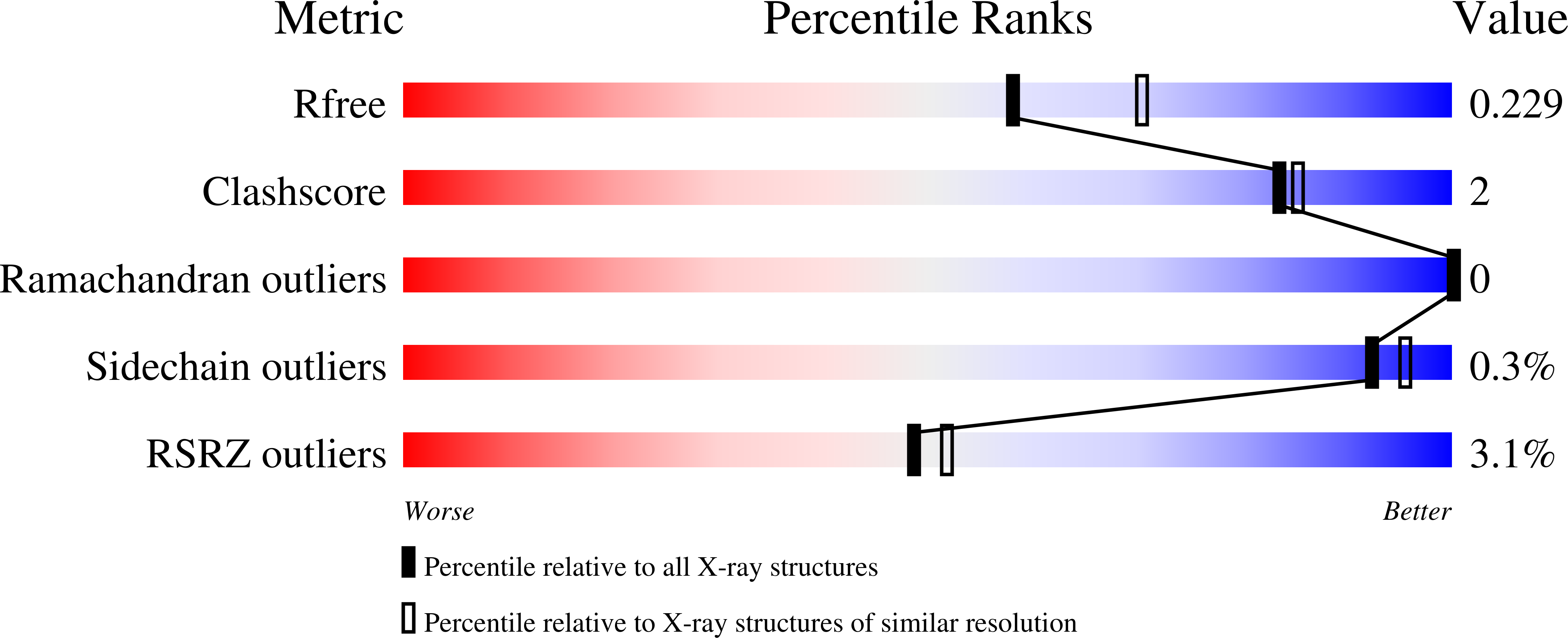
Resolution:
2.25 Å
R-Value Free:
0.22
R-Value Work:
0.18
R-Value Observed:
0.18
Space Group:
P 43 2 2