Deposition Date
2021-09-02
Release Date
2022-09-14
Last Version Date
2024-05-01
Entry Detail
PDB ID:
7PMM
Keywords:
Title:
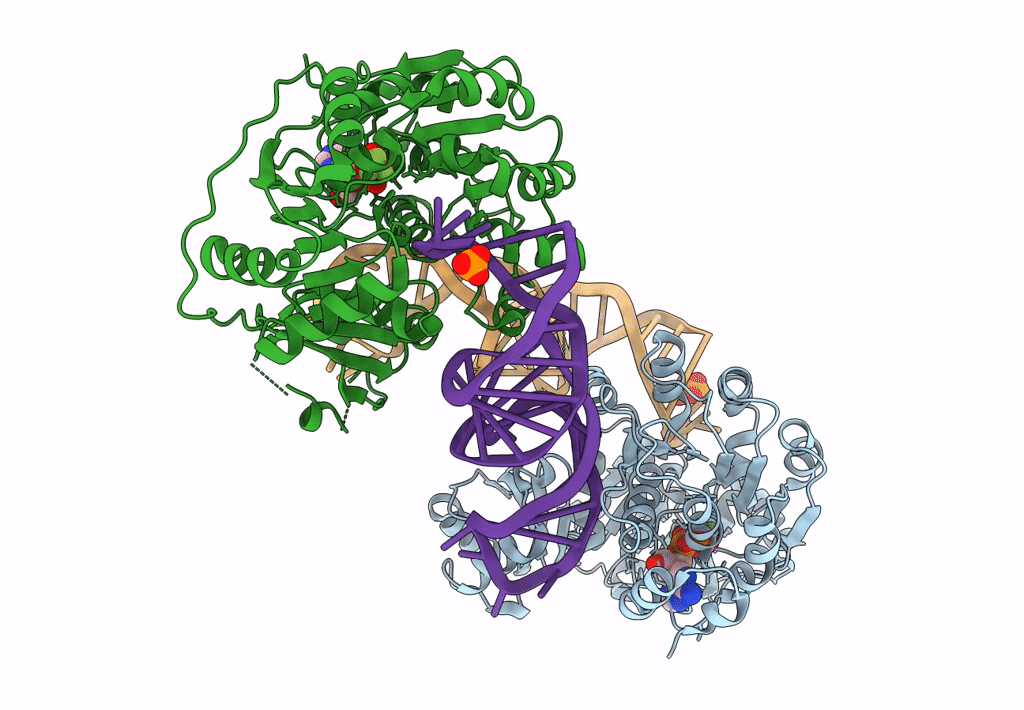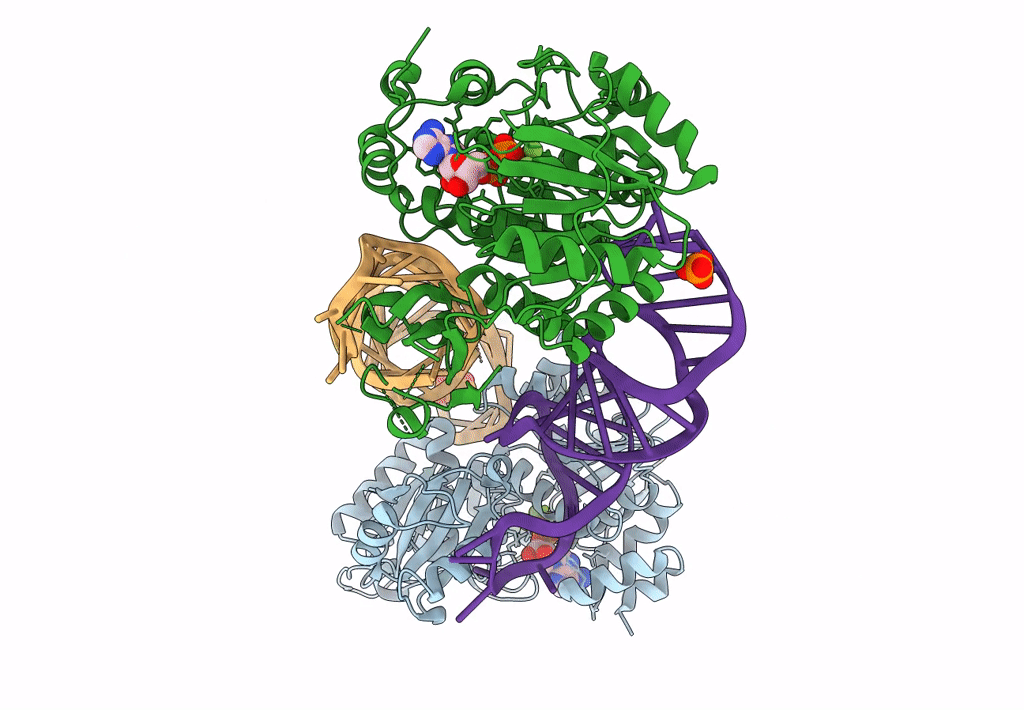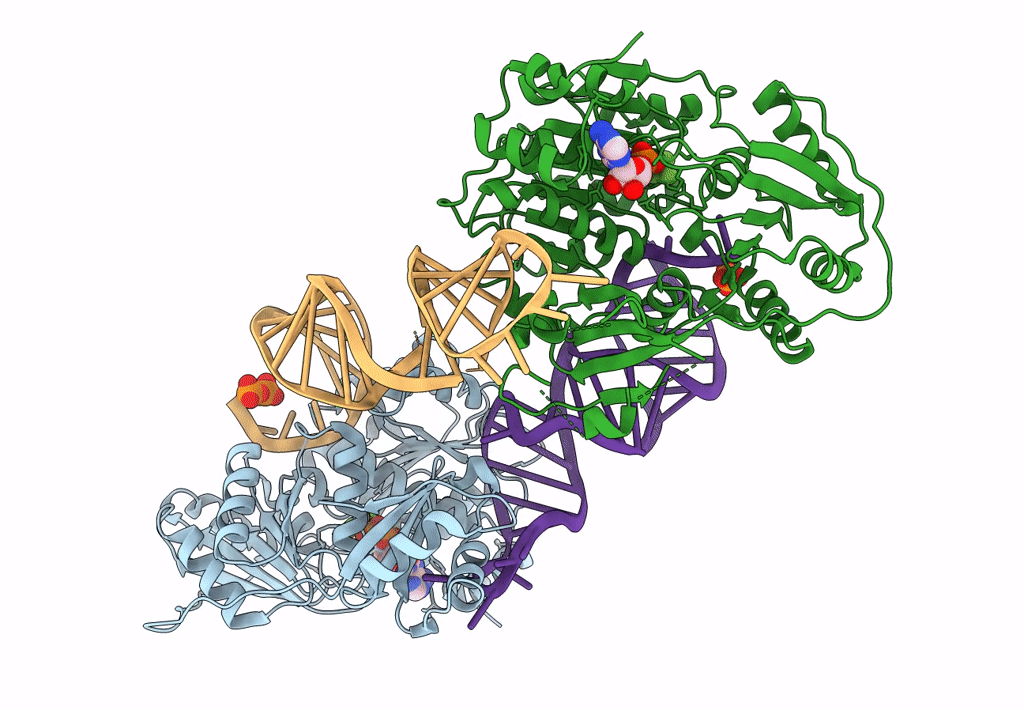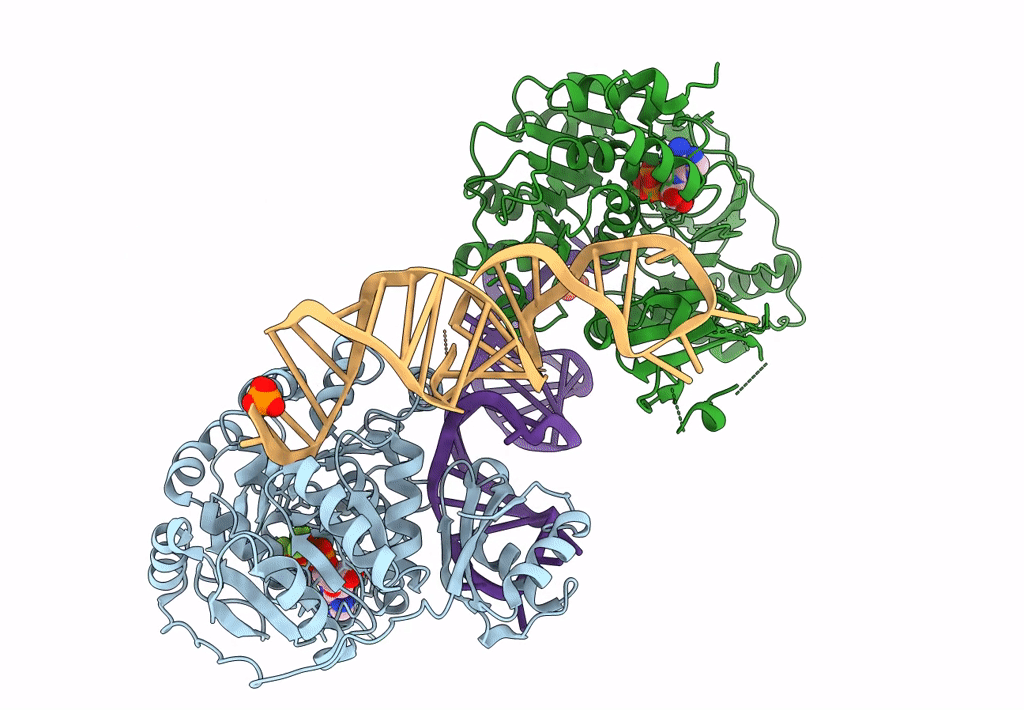
DEAD-box helicase DbpA in the active conformation bound to a ss/dsRNA junction and ADP/BeF3
Biological Source:
Source Organism:
Escherichia coli (strain K12) (Taxon ID: 83333)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
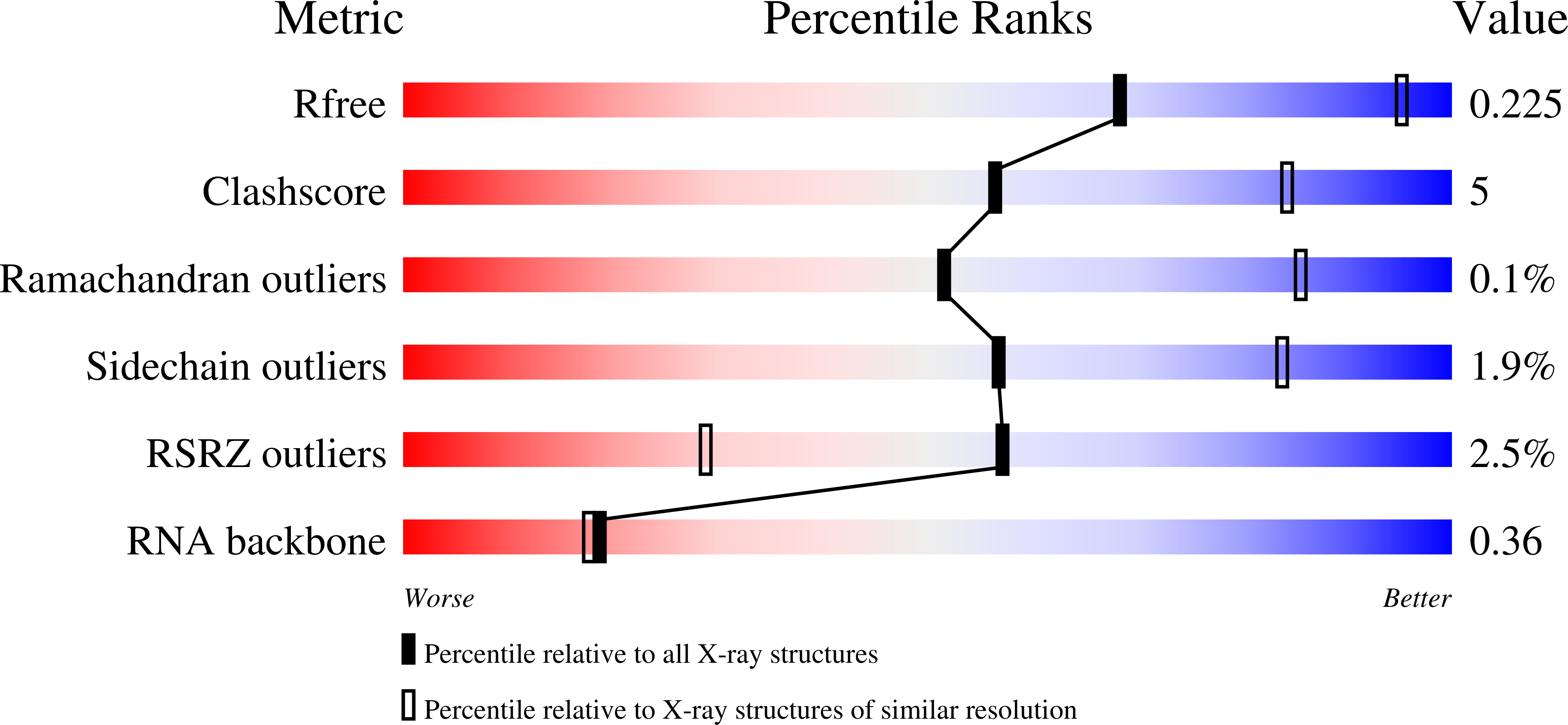
Resolution:
3.00 Å
R-Value Free:
0.22
R-Value Work:
0.19
R-Value Observed:
0.19
Space Group:
P 42 21 2