Deposition Date
2021-08-02
Release Date
2022-09-14
Last Version Date
2024-07-17
Entry Detail
PDB ID:
7PBT
Keywords:
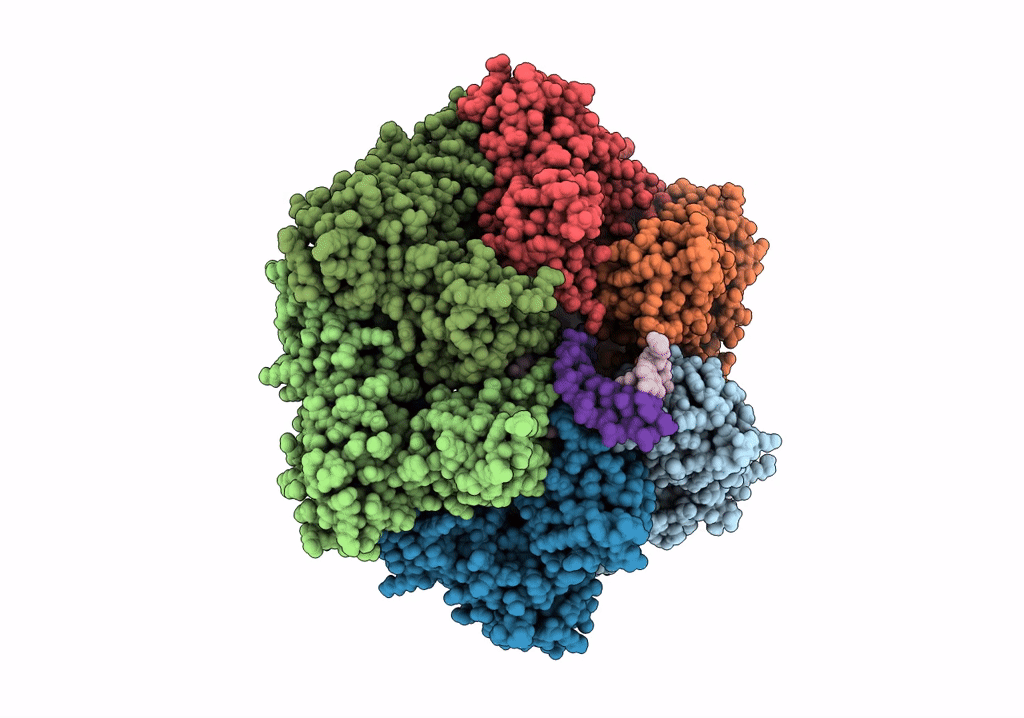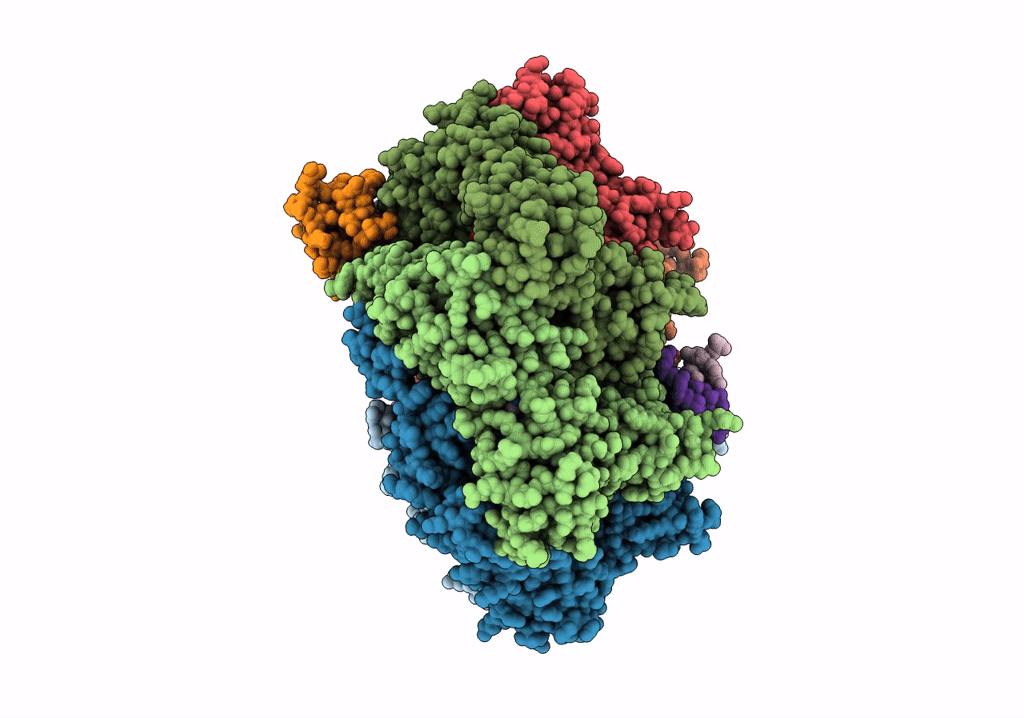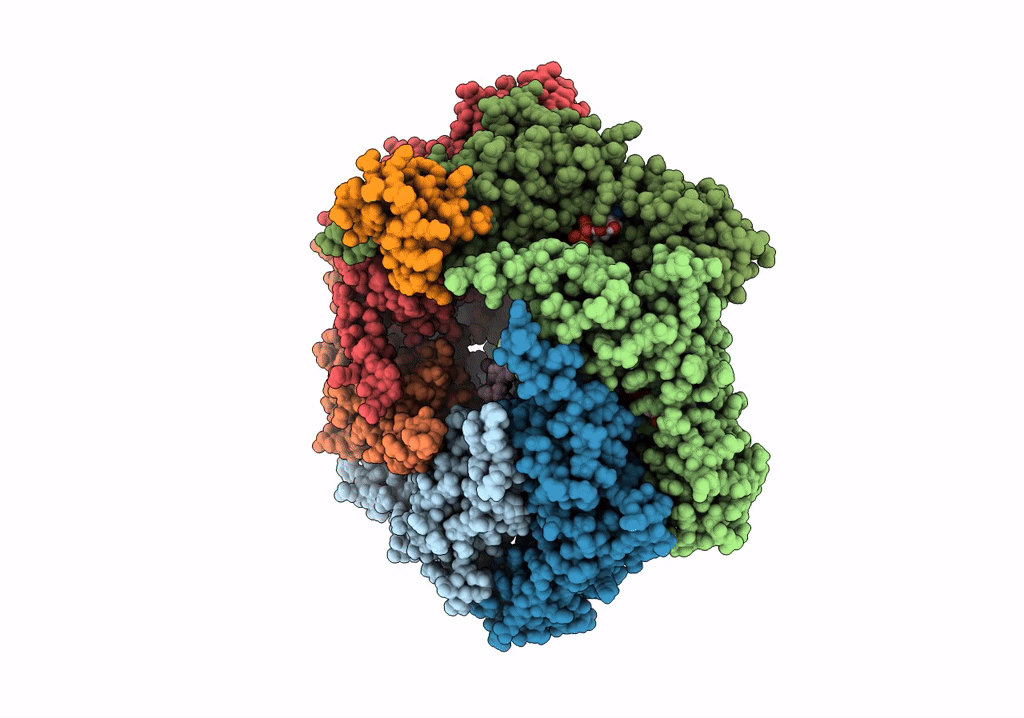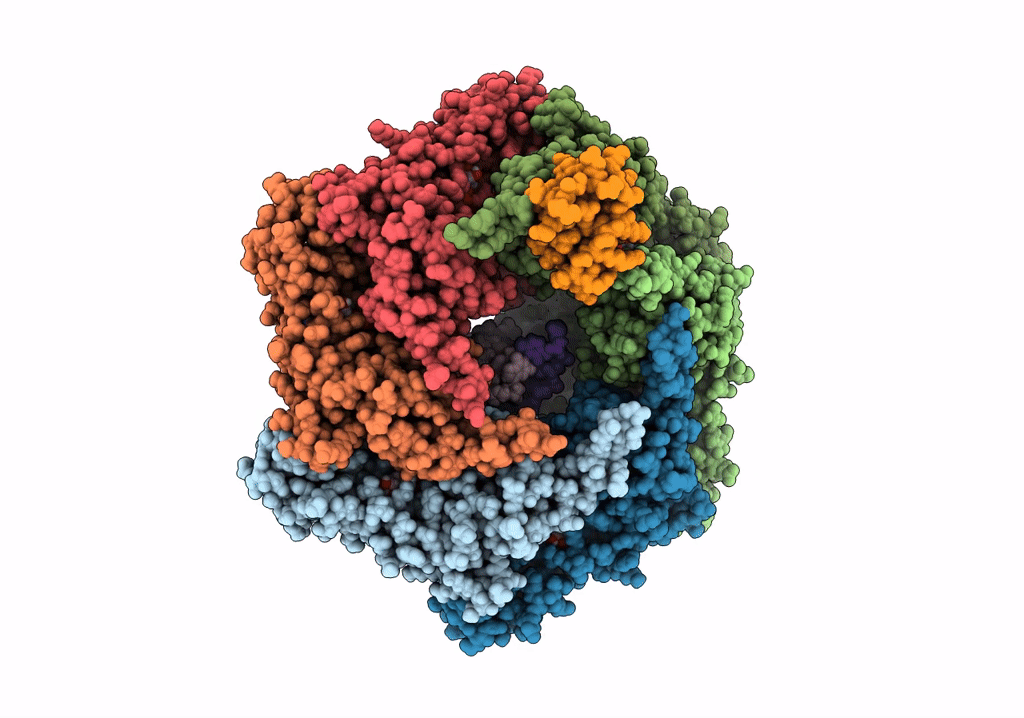
Title:
RuvAB branch migration motor complexed to the Holliday junction - RuvB AAA+ state s1 [t1 dataset]
Biological Source:
Source Organism(s):
Streptococcus thermophilus (Taxon ID: 1308)
Salmonella typhimurium (Taxon ID: 90371)
synthetic construct (Taxon ID: 32630)
Salmonella typhimurium (Taxon ID: 90371)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.30 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE


