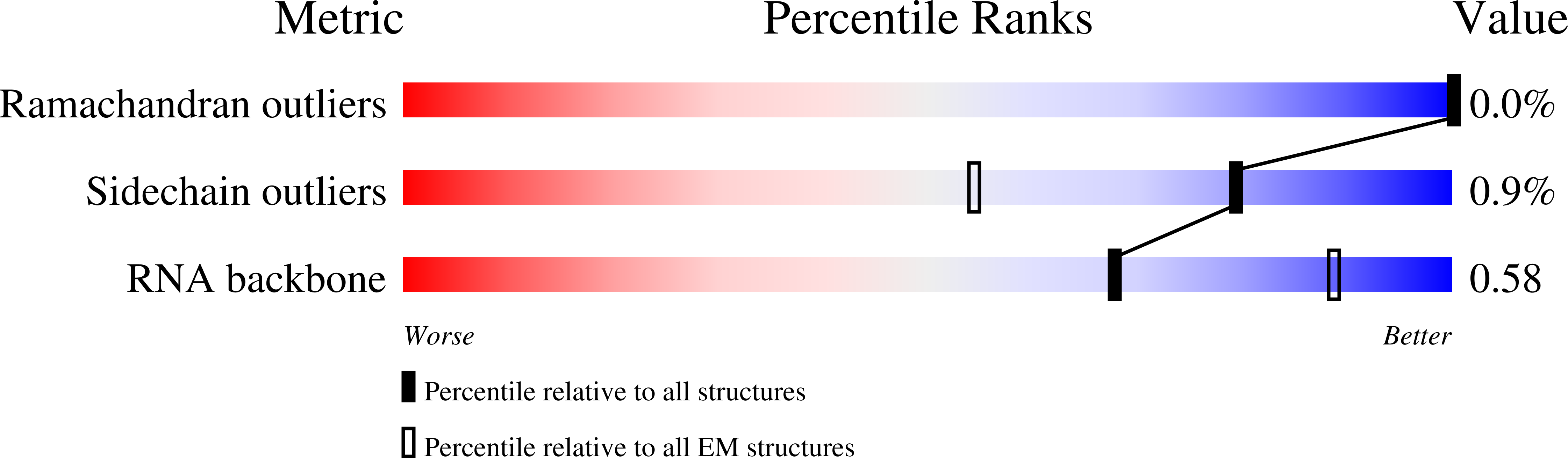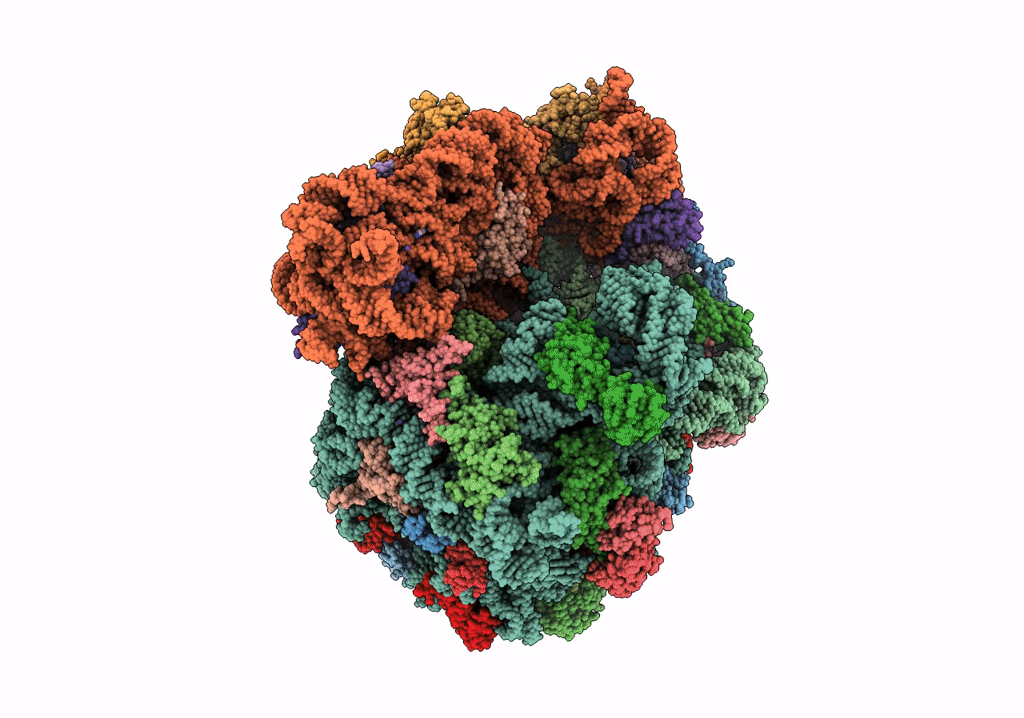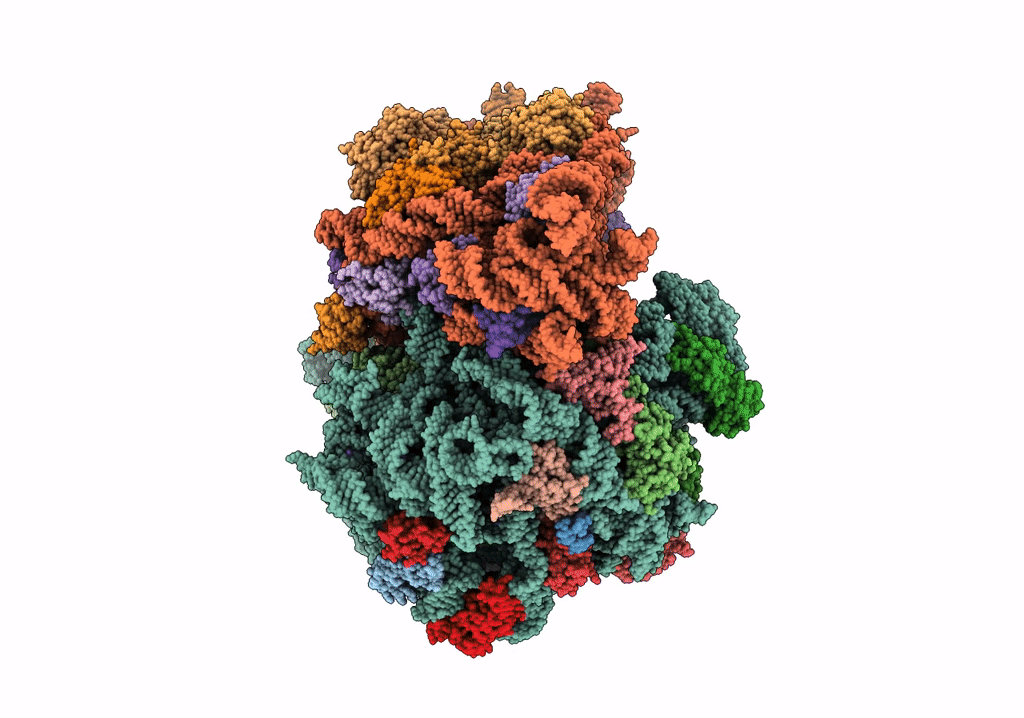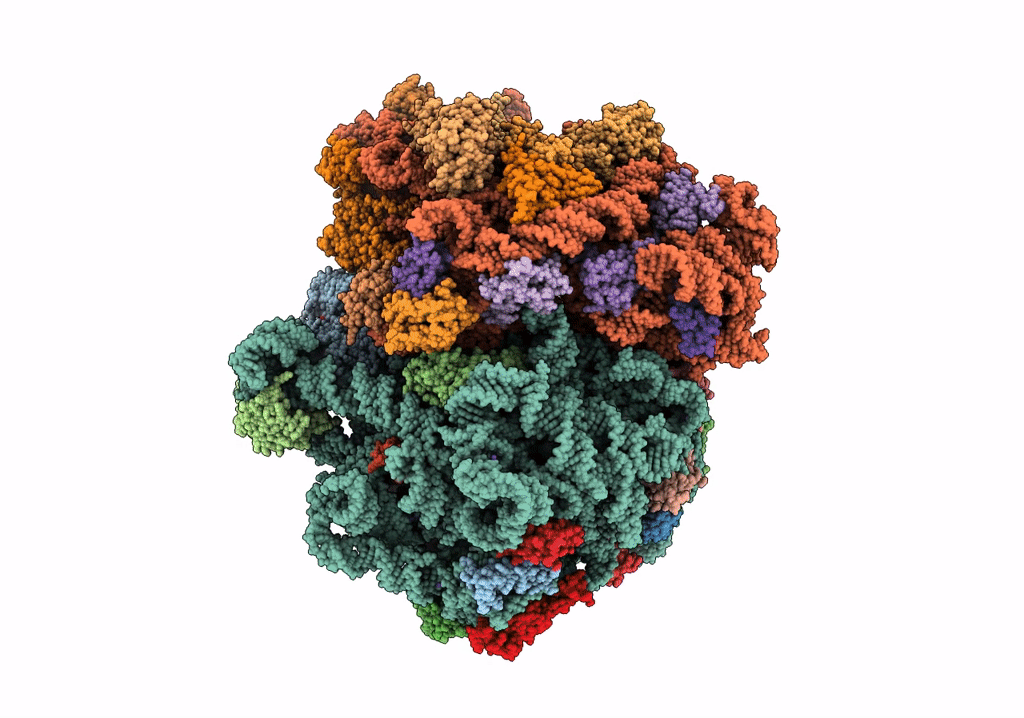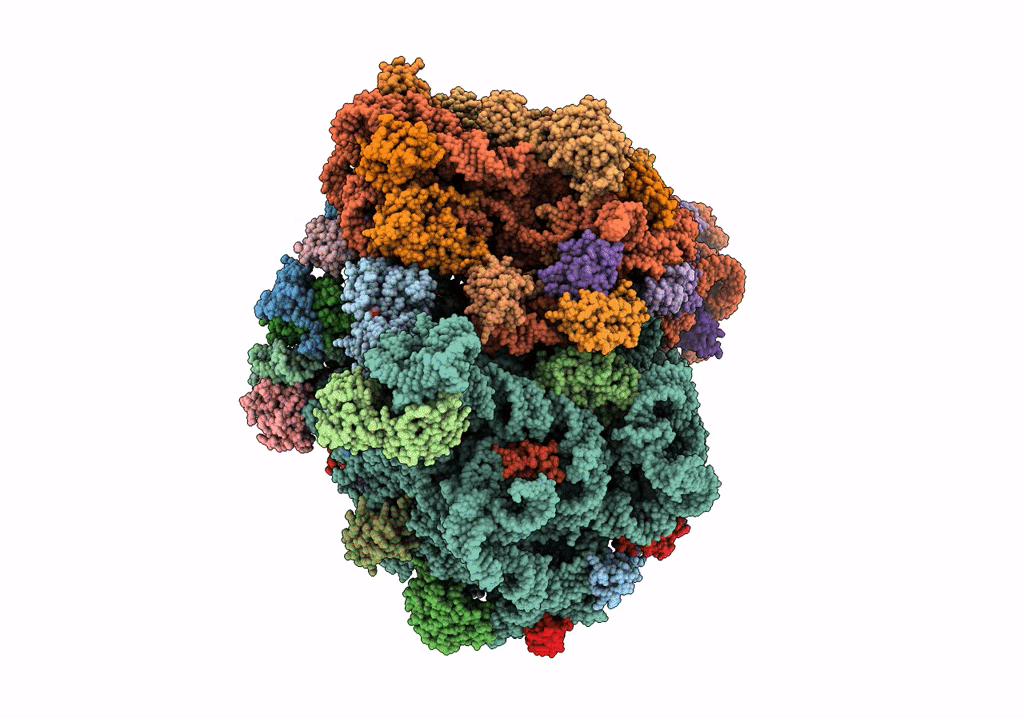
Deposition Date
2021-07-20
Release Date
2022-03-23
Last Version Date
2024-07-17
Entry Detail
PDB ID:
7P7R
Keywords:
Title:
PoxtA-EQ2 antibiotic resistance ABCF bound to E. faecalis 70S ribosome, state I
Biological Source:
Source Organism(s):
Enterococcus faecalis V583 (Taxon ID: 226185)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
2.90 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE