Deposition Date
2021-07-19
Release Date
2021-10-27
Last Version Date
2024-01-31
Entry Detail
PDB ID:
7P75
Keywords:
Title:
Re-engineered 2-deoxy-D-ribose-5-phosphate aldolase catalysing asymmetric Michael addition reactions in substrate-free state
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
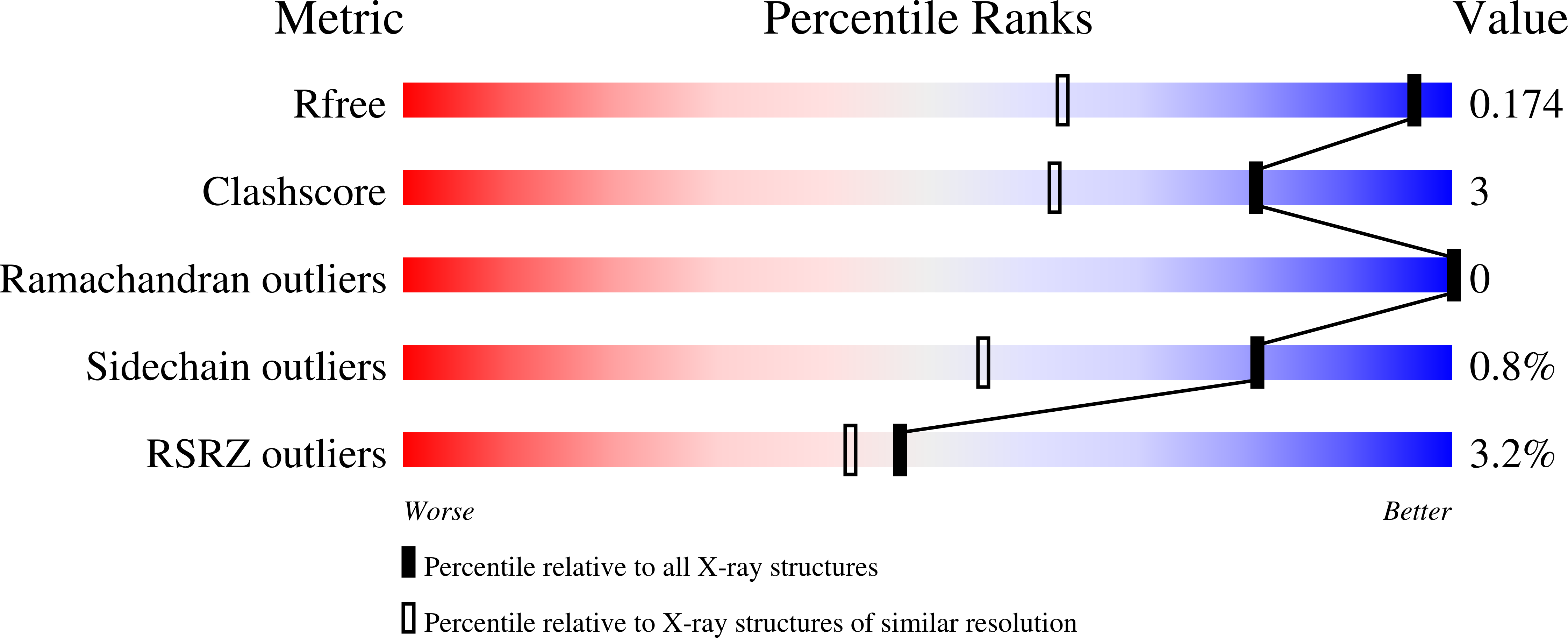
Resolution:
1.23 Å
R-Value Free:
0.17
R-Value Work:
0.14
Space Group:
P 1 21 1