Deposition Date
2021-06-28
Release Date
2021-08-18
Last Version Date
2024-07-17
Entry Detail
PDB ID:
7OZV
Keywords:
Title:
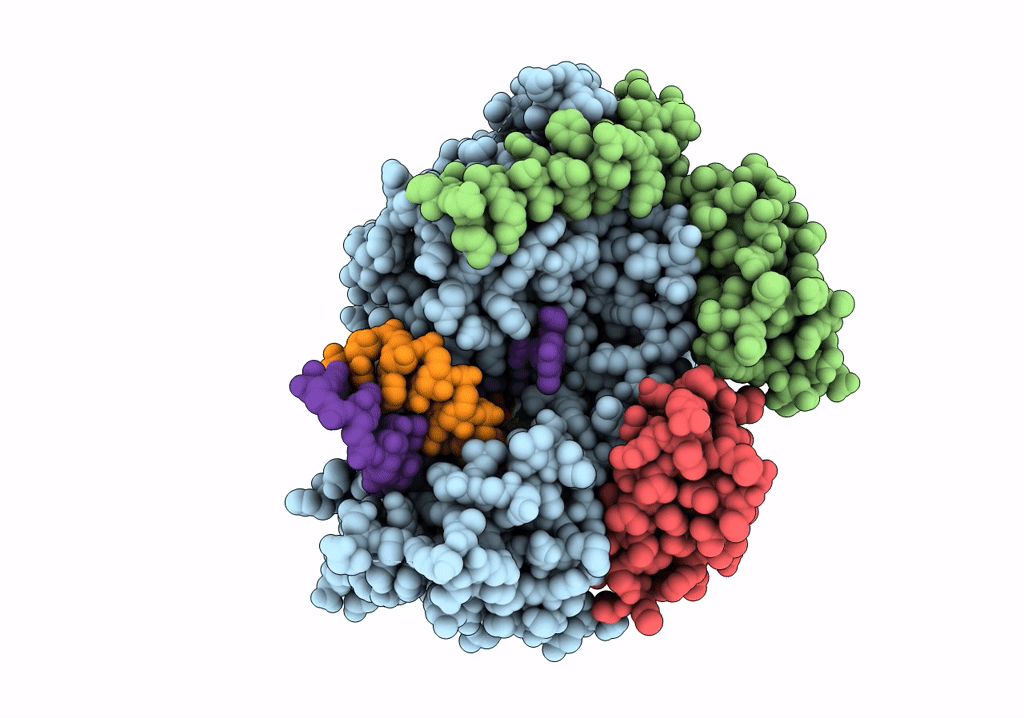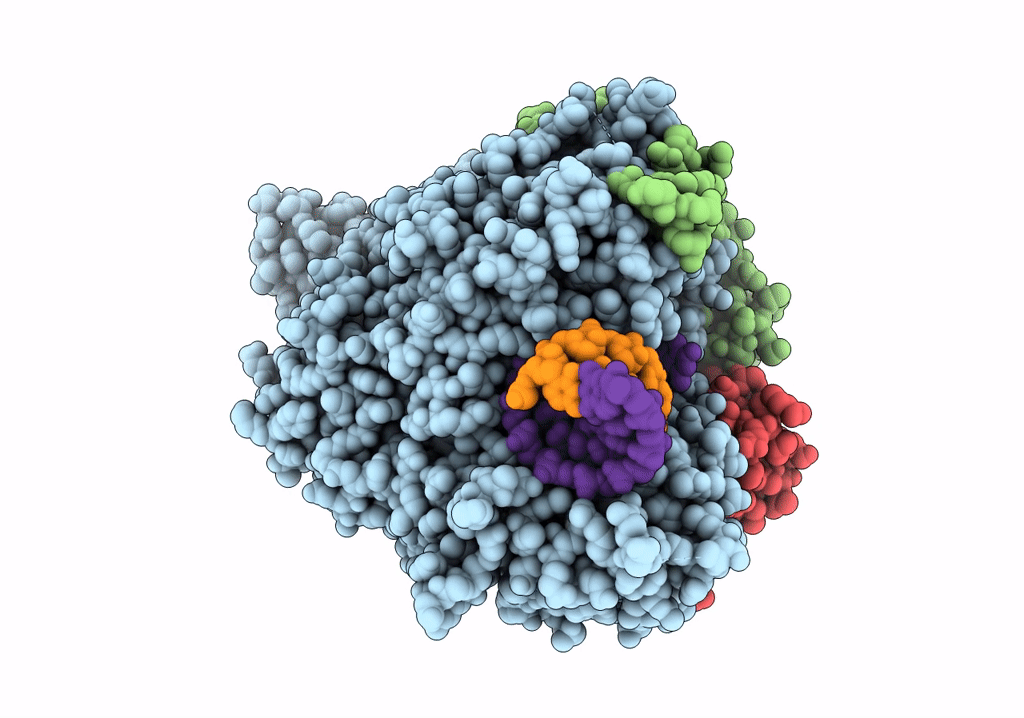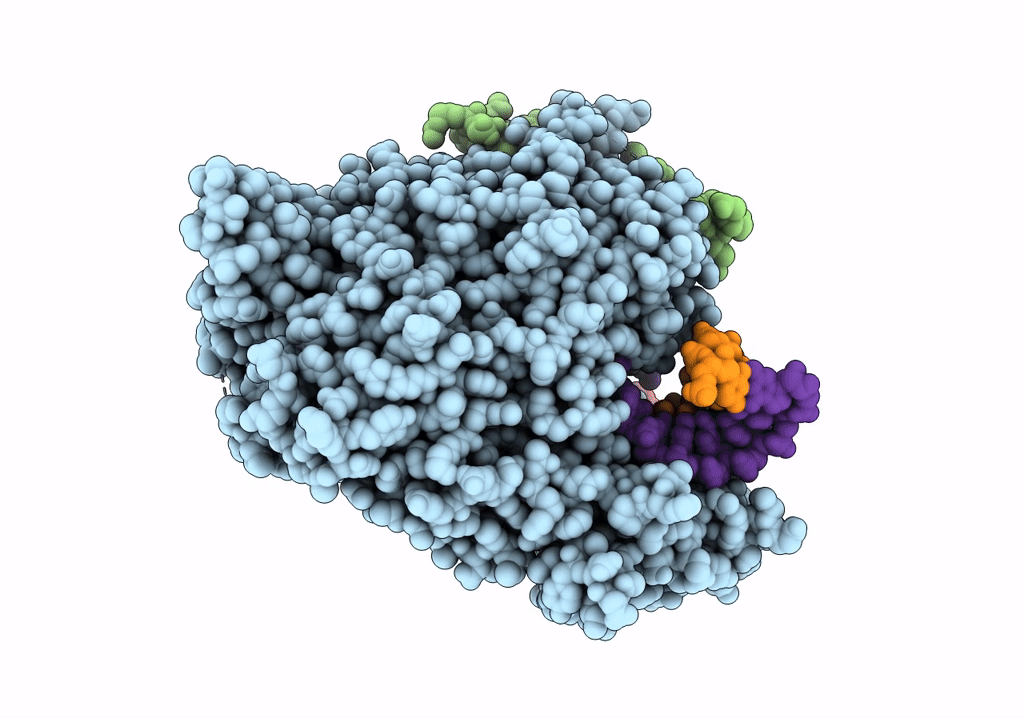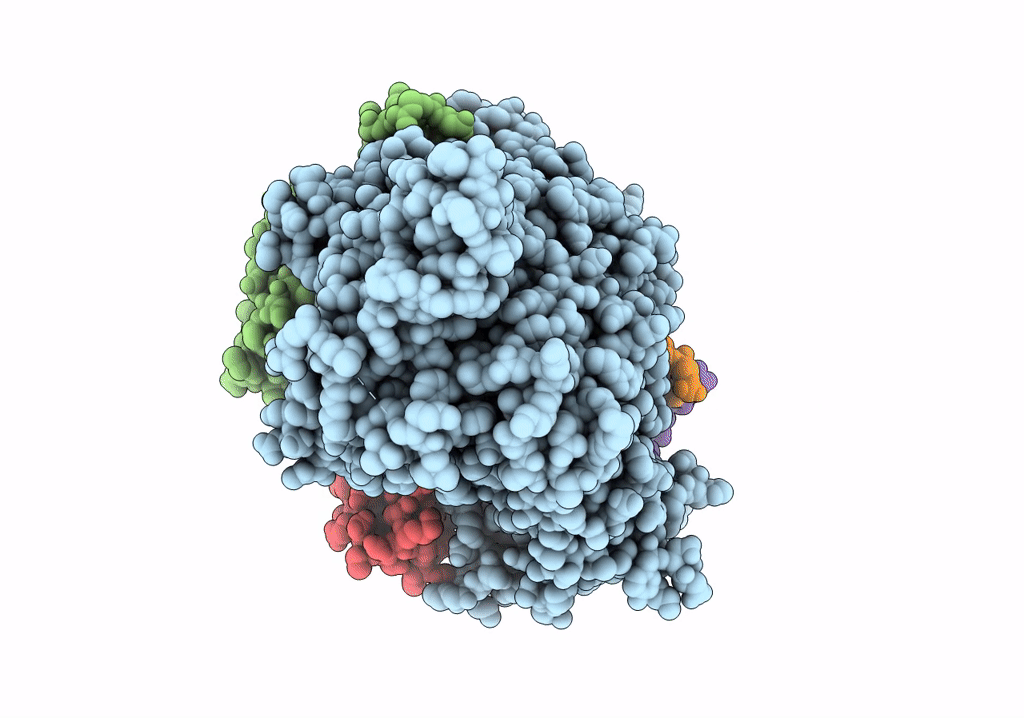
SARS-CoV-2 RdRp with Molnupiravir/ NHC in the template strand base-paired with G
Biological Source:
Source Organism:
Severe acute respiratory syndrome coronavirus 2 (Taxon ID: 2697049)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Host Organism:
Method Details:
Experimental Method:
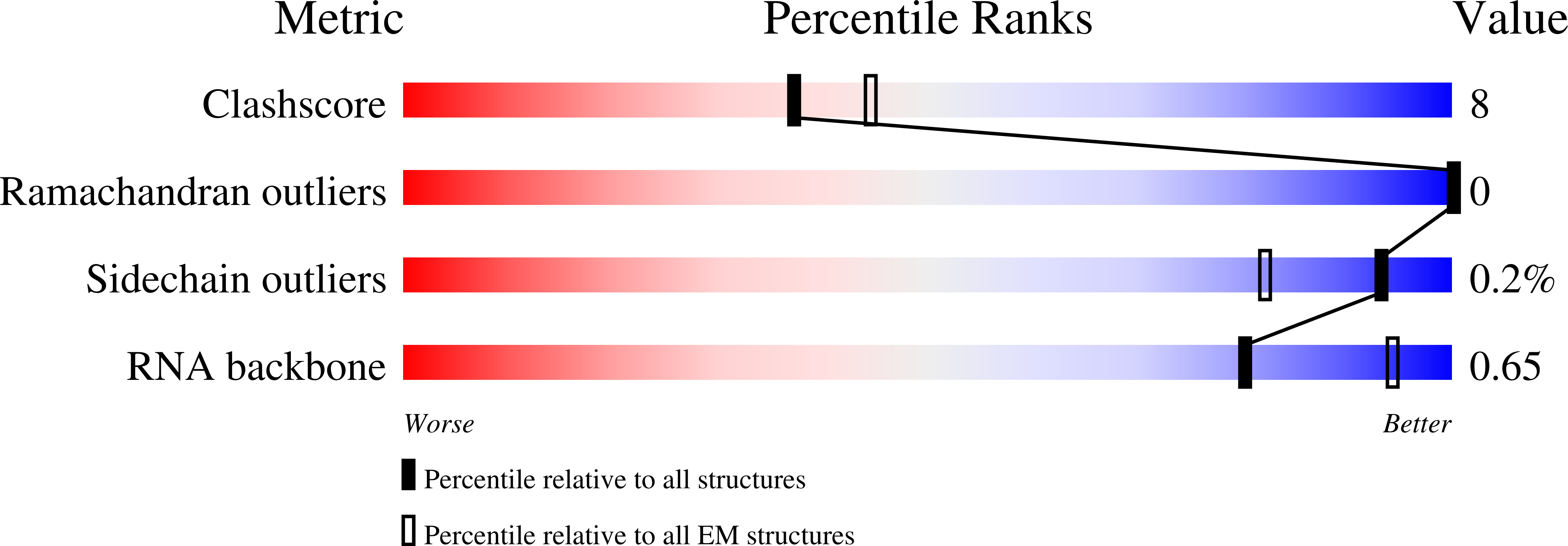
Resolution:
3.20 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE