Deposition Date
2021-06-11
Release Date
2022-06-22
Last Version Date
2024-10-09
Entry Detail
PDB ID:
7OUE
Keywords:
Title:
Crystal structure of a trapped Pab-AGOG/single-standed DNA covalent intermediate
Biological Source:
Source Organism(s):
Pyrococcus abyssi (strain GE5 / Orsay) (Taxon ID: 272844)
synthetic construct (Taxon ID: 32630)
synthetic construct (Taxon ID: 32630)
Expression System(s):
Method Details:
Experimental Method:
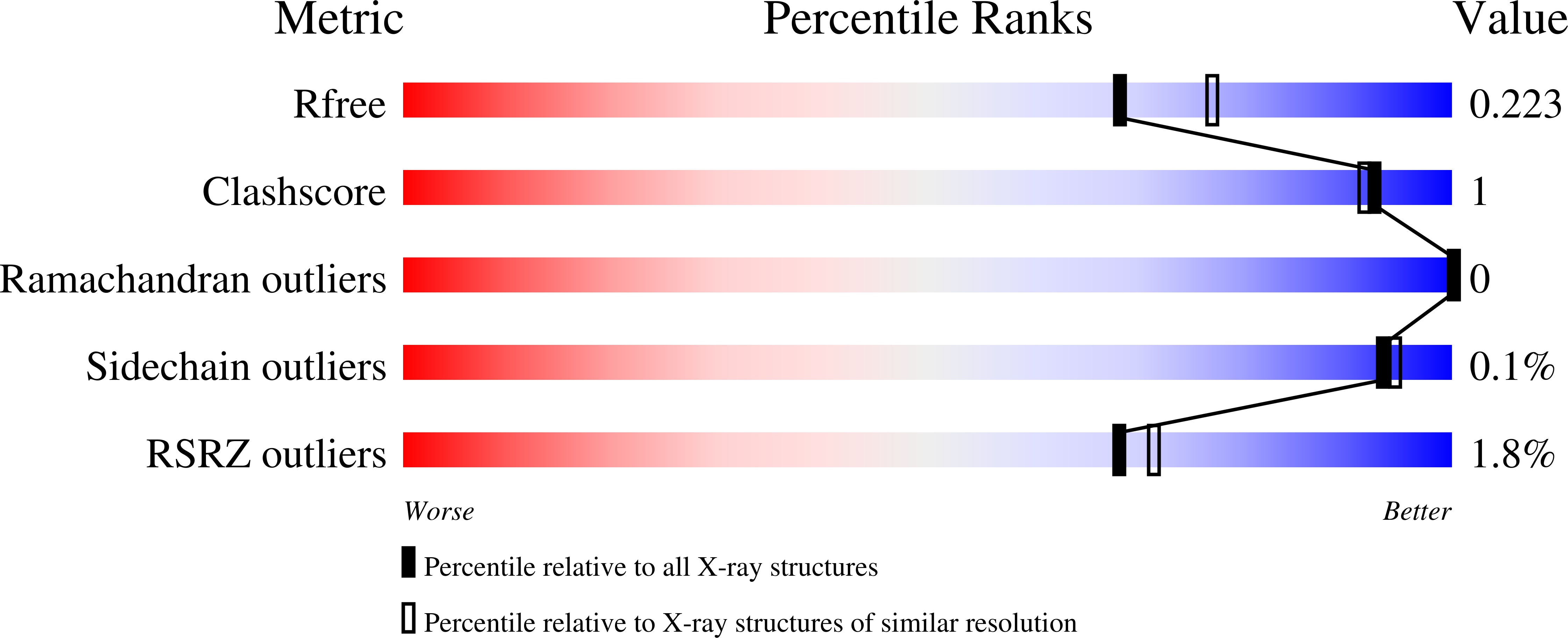
Resolution:
2.04 Å
R-Value Free:
0.22
R-Value Work:
0.17
R-Value Observed:
0.17
Space Group:
P 1