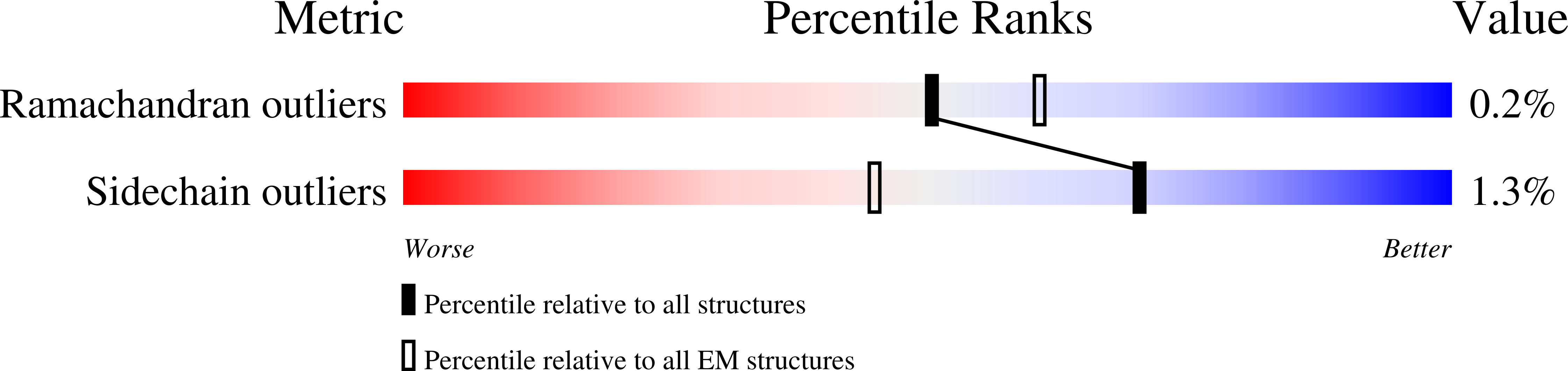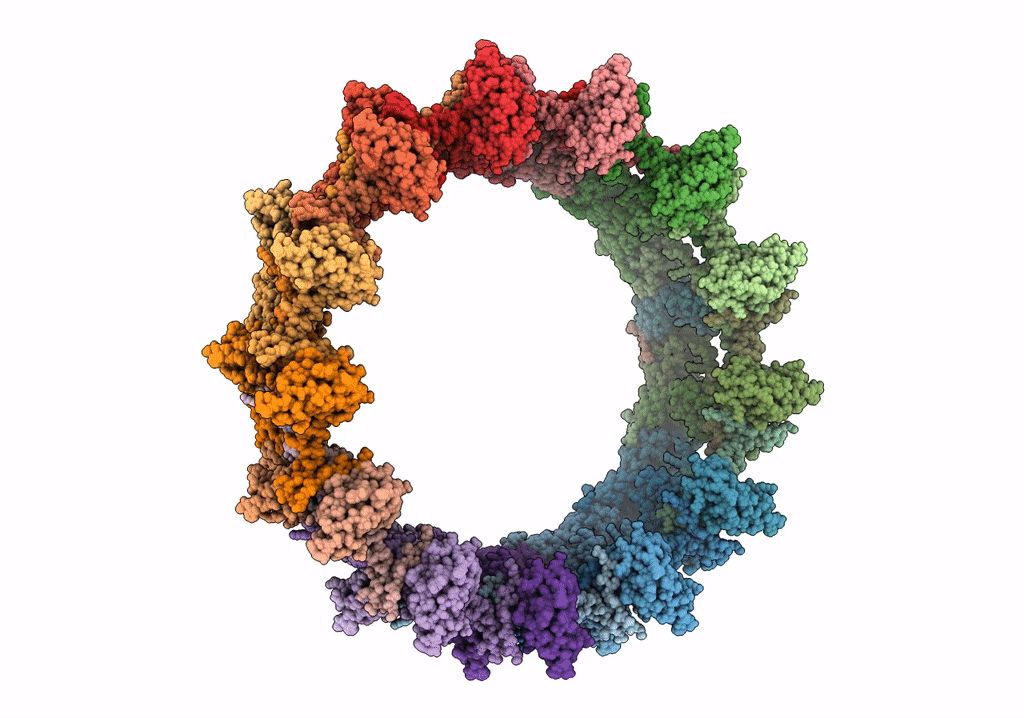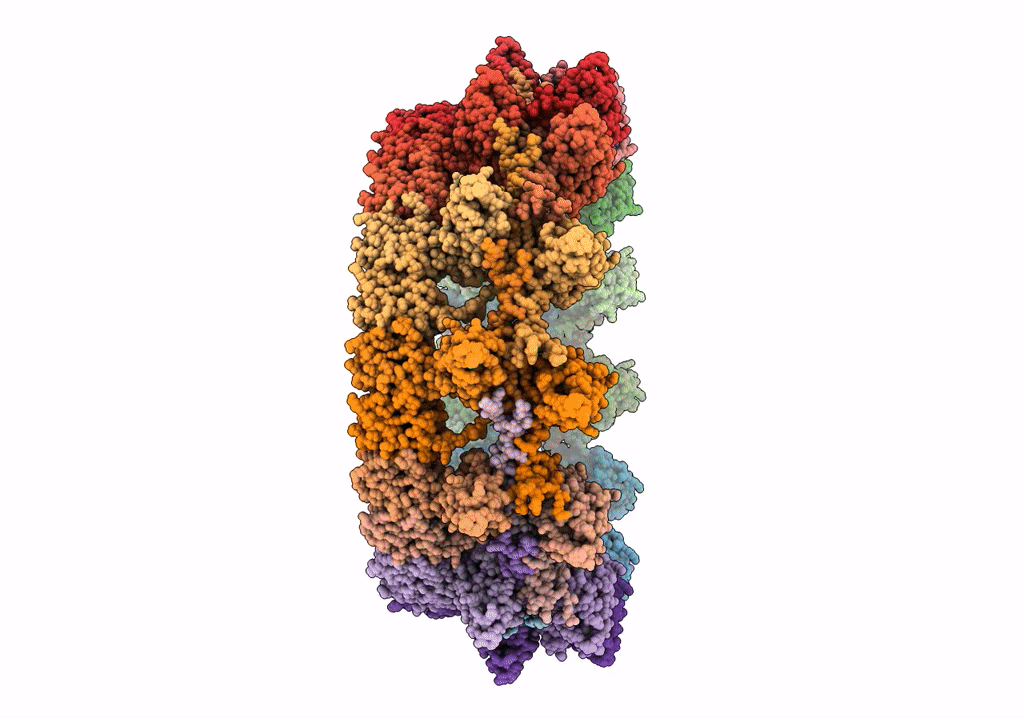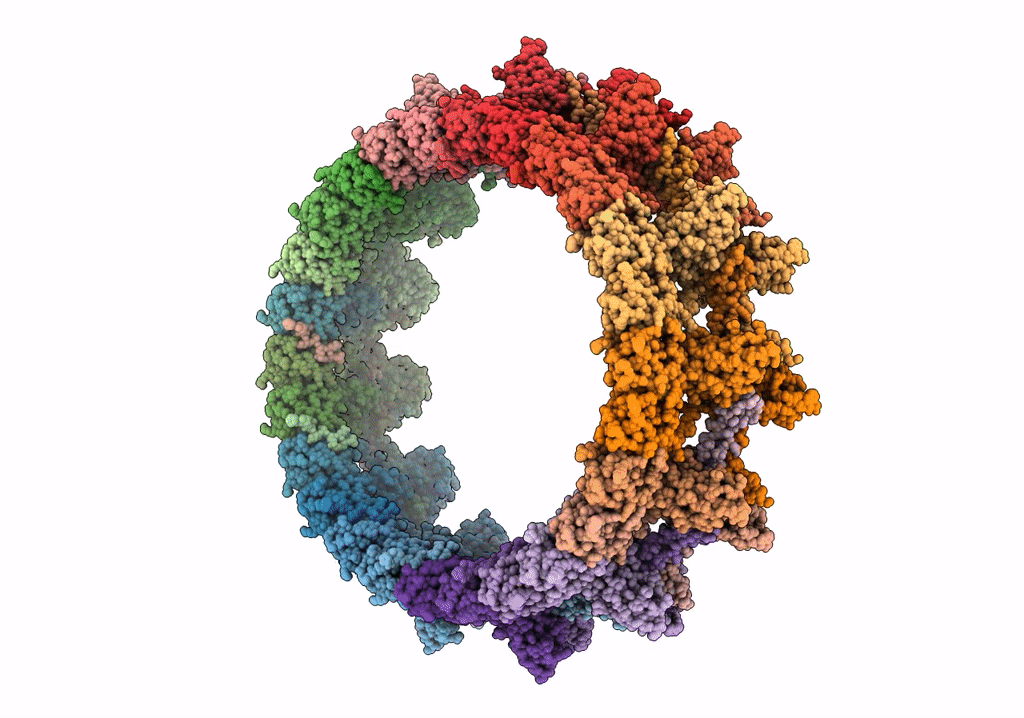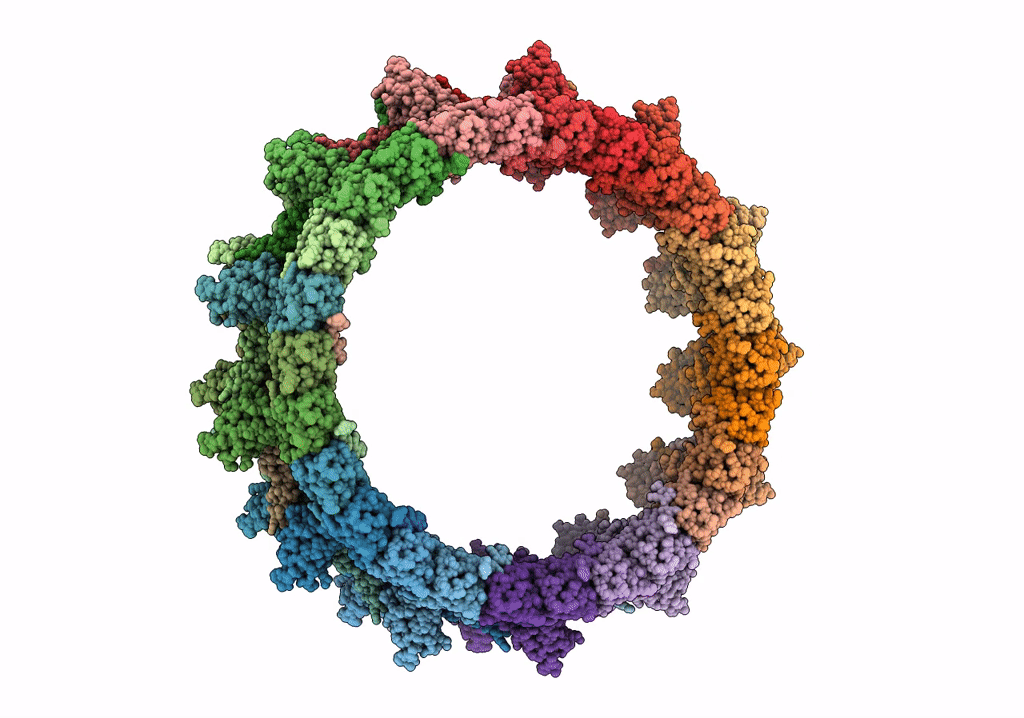
Deposition Date
2021-05-18
Release Date
2021-12-08
Last Version Date
2024-07-10
Entry Detail
PDB ID:
7OKO
Keywords:
Title:
Structure of the outer-membrane core complex (outer ring) from a conjugative type IV secretion system
Biological Source:
Source Organism(s):
Salmonella enterica (Taxon ID: 28901)
Salmonella enterica subsp. salamae serovar 58:l,z13,z28:z6 (Taxon ID: 1160778)
Salmonella enterica subsp. salamae serovar 58:l,z13,z28:z6 (Taxon ID: 1160778)
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.40 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE