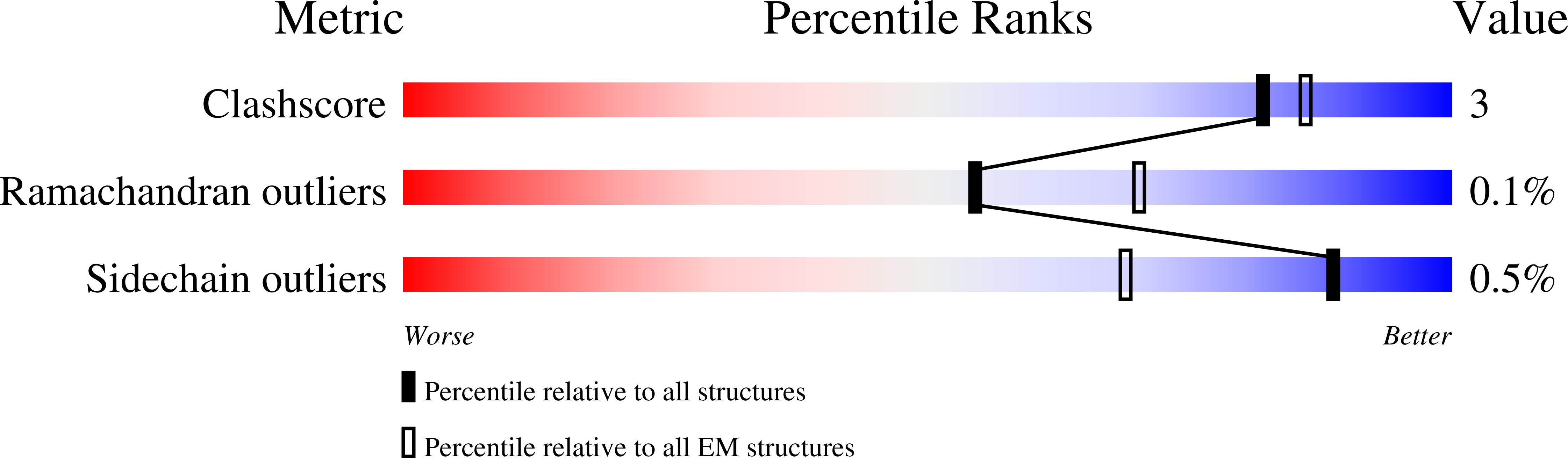
Deposition Date
2021-04-29
Release Date
2022-01-26
Last Version Date
2024-11-13
Entry Detail
PDB ID:
7ODL
Keywords:
Title:
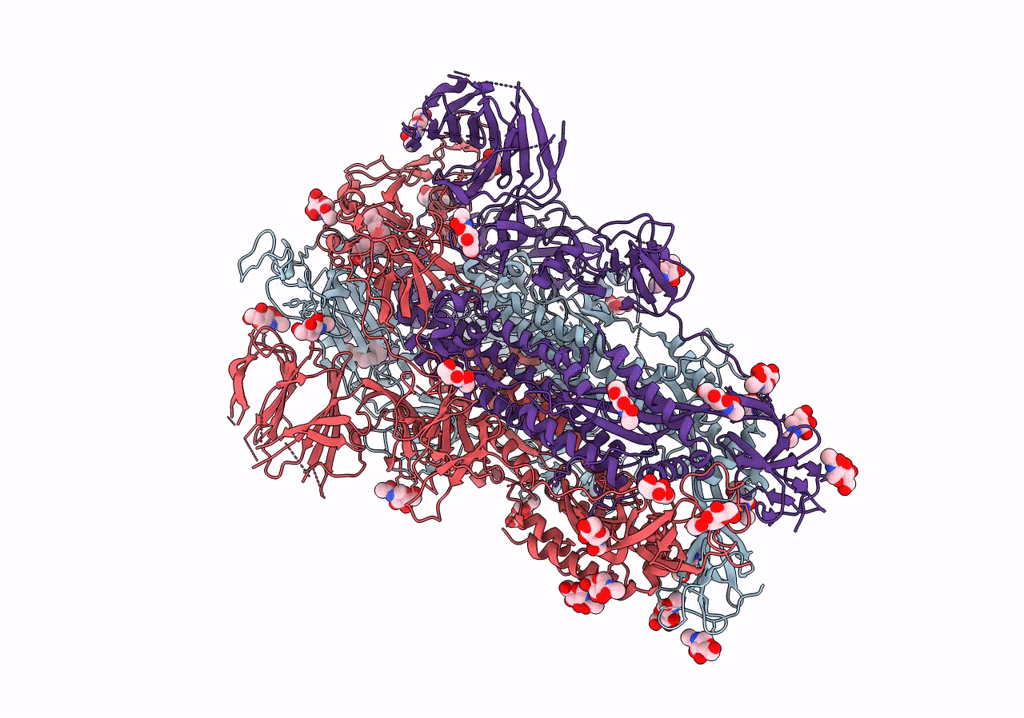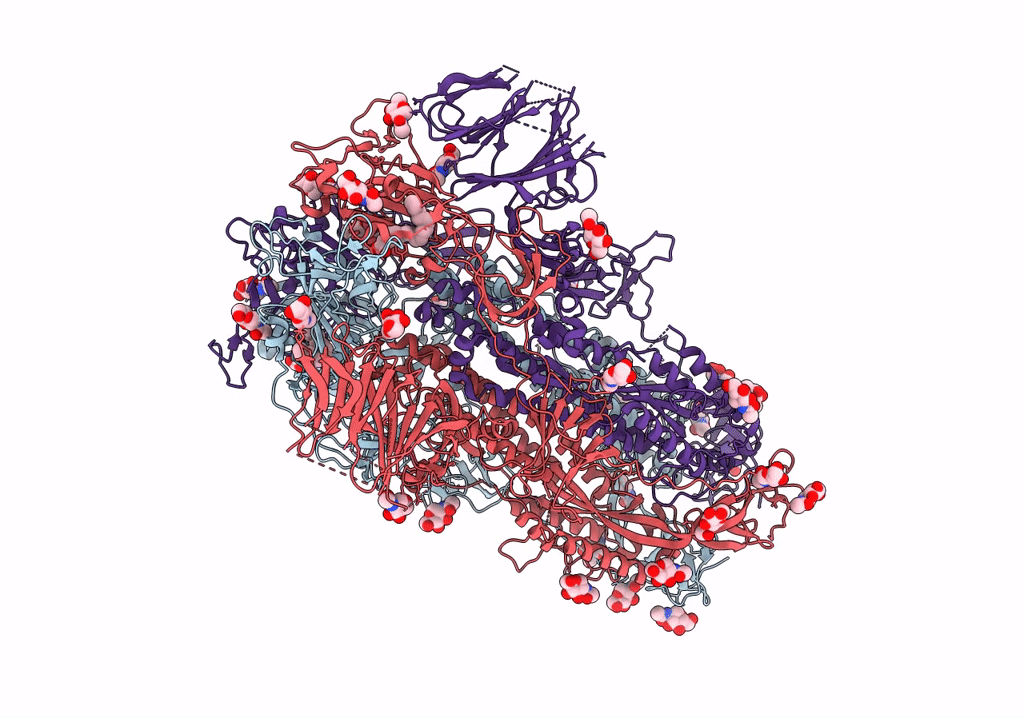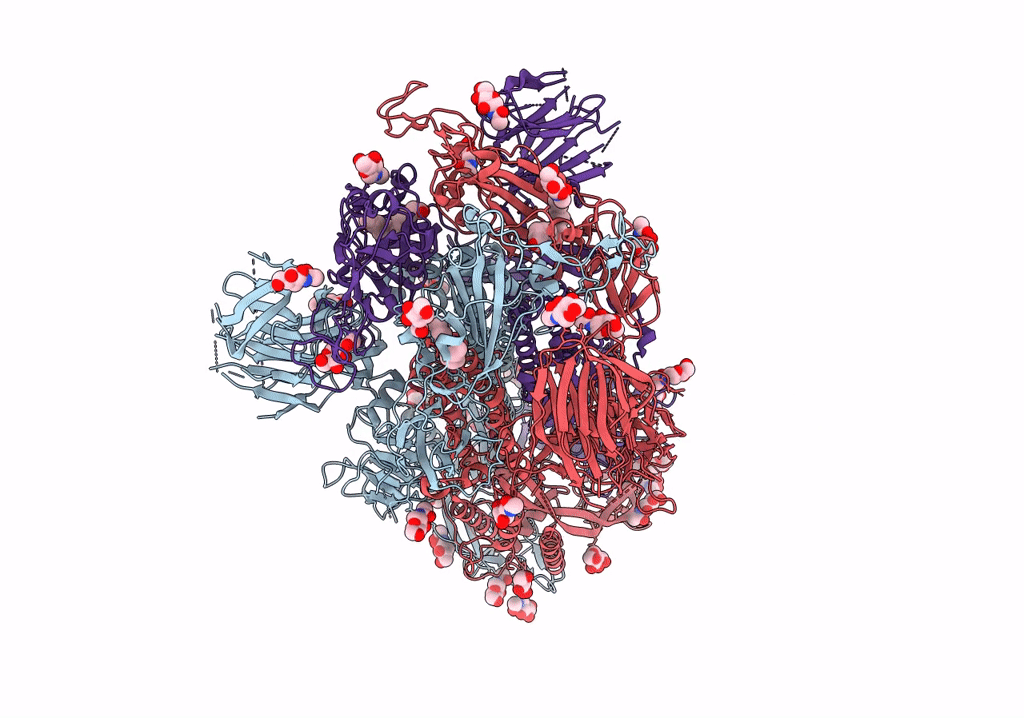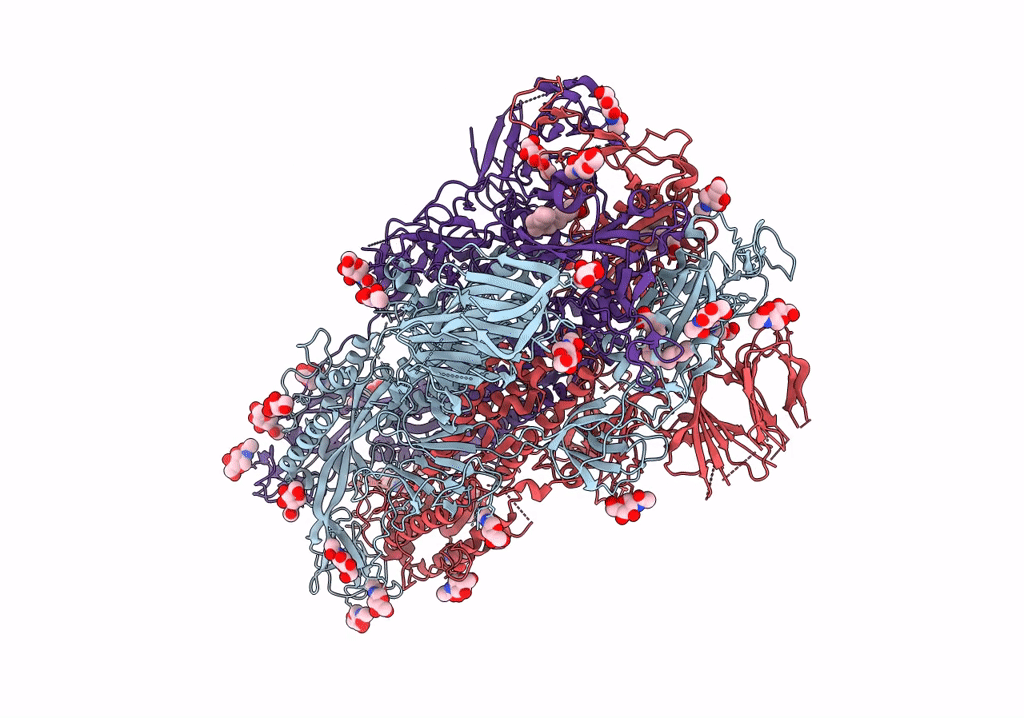
SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry
Biological Source:
Source Organism(s):
Expression System(s):
Method Details:
Experimental Method:
Resolution:
3.03 Å
Aggregation State:
PARTICLE
Reconstruction Method:
SINGLE PARTICLE